Long noncoding RNA Regulating ImMune Escape regulates mixed lineage leukaemia protein-1-H3K4me3-mediated immune escape in oesophageal squamous cell carcinoma
- PMID: 37712124
- PMCID: PMC10502462
- DOI: 10.1002/ctm2.1410
Long noncoding RNA Regulating ImMune Escape regulates mixed lineage leukaemia protein-1-H3K4me3-mediated immune escape in oesophageal squamous cell carcinoma
Abstract
Background: Predictive biomarkers for oesophageal squamous cell carcinoma (ESCC) immunotherapy are lacking, and immunotherapy resistance remains to be addressed. The role of long noncoding RNA (lncRNA) in ESCC immune escape and immunotherapy resistance remains to be elucidated.
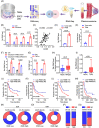
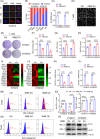
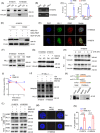
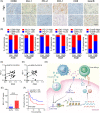
Methods: The tumour-associated macrophage-upregulated lncRNAs and the exosomal lncRNAs highly expressed in ESCC immunotherapy nonresponders were identified by lncRNA sequencing and polymerase chain reaction assays. CRISPR-Cas9 was used to explore the functional roles of the lncRNA. RNA pull-down, MS2-tagged RNA affinity purification (MS2-TRAP) and RNA-binding protein immunoprecipitation (RIP) were performed to identify lncRNA-associated proteins and related mechanisms. In vivo, the humanized PBMC (hu-PBMC) mouse model was established to assess the therapeutic responses of specific lncRNA inhibitors and their combination with programmed cell death protein 1 (PD-1) monoclonal antibody (mAb). Single-cell sequencing, flow cytometry, and multiplex fluorescent immunohistochemistry were used to analyze immune cells infiltrating the tumour microenvironment.
Results: We identified a lncRNA that is involved in tumour immune evasion and immunotherapy resistance. High LINC02096 (RIME) expression in plasma exosomes correlates with a reduced response to PD-1 mAb treatment and poor prognosis. Mechanistically, RIME binds to mixed lineage leukaemia protein-1 (MLL1) and prevents ankyrin repeat and SOCS box containing 2 (ASB2)-mediated MLL1 ubiquitination, improving the stability of MLL1. RIME-MLL1 increases H3K4me3 levels in the promoter regions of programmed death-ligand 1 (PD-L1) and indoleamine 2,3-dioxygenase 1 (IDO-1), constitutively increasing the expression of PD-L1/IDO-1 in tumour cells and inhibiting CD8+ T cells infiltration and activation. RIME depletion in huPBMC-NOG mice significantly represses tumour development and improves the effectiveness of PD-1 mAb treatment by activating T-cell-mediated antitumour immunity.
Conclusions: This study reveals that the RIME-MLL1-H3K4me3 axis plays a critical role in tumour immunosuppression. Moreover, RIME appears to be a potential prognostic biomarker for immunotherapy and developing drugs that target RIME may be a new therapeutic strategy that overcomes immunotherapy resistance and benefits patients with ESCC.
Keywords: esophageal squamous cell carcinoma; immune escape; immunotherapy; lncRNA; mixed lineage leukaemia protein-1 (MLL1).
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion.J Natl Cancer Inst. 2017 Jan 28;109(6):djw283. doi: 10.1093/jnci/djw283. Print 2017 Jan. J Natl Cancer Inst. 2017. PMID: 28131992 Free PMC article.
-
CircNF1 modulates the progression and immune evasion of esophageal squamous cell carcinoma through dual regulation of PD-L1.Cell Mol Biol Lett. 2025 Mar 29;30(1):37. doi: 10.1186/s11658-025-00712-y. Cell Mol Biol Lett. 2025. PMID: 40158127 Free PMC article.
-
Molecular mechanism of lncRNA SNHG12 in immune escape of non-small cell lung cancer through the HuR/PD-L1/USP8 axis.Cell Mol Biol Lett. 2022 Jun 3;27(1):43. doi: 10.1186/s11658-022-00343-7. Cell Mol Biol Lett. 2022. PMID: 35658874 Free PMC article.
-
Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma.Cancer Sci. 2020 Sep;111(9):3132-3141. doi: 10.1111/cas.14541. Epub 2020 Jul 14. Cancer Sci. 2020. PMID: 32579769 Free PMC article. Review.
-
Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure).Ann Oncol. 2016 Aug;27(8):1492-504. doi: 10.1093/annonc/mdw217. Epub 2016 May 20. Ann Oncol. 2016. PMID: 27207108 Review.
Cited by
-
Latest insights into the global epidemiological features, screening, early diagnosis and prognosis prediction of esophageal squamous cell carcinoma.World J Gastroenterol. 2024 May 28;30(20):2638-2656. doi: 10.3748/wjg.v30.i20.2638. World J Gastroenterol. 2024. PMID: 38855150 Free PMC article. Review.
-
Personalized mRNA vaccine combined with PD-1 inhibitor therapy in a patient with advanced esophageal squamous cell carcinoma.Am J Cancer Res. 2024 Aug 25;14(8):3896-3904. doi: 10.62347/NVFB3780. eCollection 2024. Am J Cancer Res. 2024. PMID: 39267685 Free PMC article.
-
Helicobacter pylori infection delays neutrophil apoptosis and exacerbates inflammatory response.Future Microbiol. 2024 Sep;19(13):1145-1156. doi: 10.1080/17460913.2024.2360798. Epub 2024 Jul 26. Future Microbiol. 2024. PMID: 39056165 Free PMC article.
-
LncRNAs in hypoxic microenvironment; insight in their impact in cancer biology.Funct Integr Genomics. 2025 Jul 2;25(1):142. doi: 10.1007/s10142-025-01635-9. Funct Integr Genomics. 2025. PMID: 40601072 Review.
-
Emerging roles of non-coding RNA derived from extracellular vesicles in regulating PD-1/PD-L1 pathway: insights into cancer immunotherapy and clinical applications.Cancer Cell Int. 2025 May 23;25(1):188. doi: 10.1186/s12935-025-03809-8. Cancer Cell Int. 2025. PMID: 40410719 Free PMC article. Review.
References
-
- Sun JM, Shen L, Shah MA, et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first‐line treatment of advanced oesophageal cancer (KEYNOTE‐590): a randomised, placebo‐controlled, phase 3 study. Lancet. 2021;398(10302):759‐771. - PubMed
-
- Wang ZX, Cui C, Yao J, et al. Toripalimab plus chemotherapy in treatment‐naïve, advanced esophageal squamous cell carcinoma (JUPITER‐06): a multi‐center phase 3 trial. Cancer Cell. 2022;40(3):277‐288. e3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
