TFEB and TFE3 control glucose homeostasis by regulating insulin gene expression
- PMID: 37712288
- PMCID: PMC10620765
- DOI: 10.15252/embj.2023113928
TFEB and TFE3 control glucose homeostasis by regulating insulin gene expression
Abstract
To fulfill their function, pancreatic beta cells require precise nutrient-sensing mechanisms that control insulin production. Transcription factor EB (TFEB) and its homolog TFE3 have emerged as crucial regulators of the adaptive response of cell metabolism to environmental cues. Here, we show that TFEB and TFE3 regulate beta-cell function and insulin gene expression in response to variations in nutrient availability. We found that nutrient deprivation in beta cells promoted TFEB/TFE3 activation, which resulted in suppression of insulin gene expression. TFEB overexpression was sufficient to inhibit insulin transcription, whereas beta cells depleted of both TFEB and TFE3 failed to suppress insulin gene expression in response to amino acid deprivation. Interestingly, ChIP-seq analysis showed binding of TFEB to super-enhancer regions that regulate insulin transcription. Conditional, beta-cell-specific, Tfeb-overexpressing, and Tfeb/Tfe3 double-KO mice showed severe alteration of insulin transcription, secretion, and glucose tolerance, indicating that TFEB and TFE3 are important physiological mediators of pancreatic function. Our findings reveal a nutrient-controlled transcriptional mechanism that regulates insulin production, thus playing a key role in glucose homeostasis at both cellular and organismal levels.
Keywords: TFEB; beta cells; glucose homeostasis; insulin; mTORC1.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
A.B. is cofounder of CASMA Therapeutics, Inc., and Advisory board member of NexGeneration Diagnostics and Avilar Therapeutics.
Figures
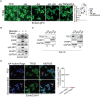
- A
Representative images of high content analyses using TFEB antibodies on fixed cells previously treated with full medium (FED), without amino acids (−aa) for 1 h, without glucose (−glu) for 1 h, without amino acids and glucose (−aa ‐glu) for 1 h or starved for 1 h and restimulated with amino acids for 30′ (−aa/Refeeding). The relative quantification (see Materials and Methods) is shown in the graph. Data are represented as mean ± standard error. Each dot represents a well from the 96‐well plate (n = 30 fed, n = 12 −aa, n = 14 −glu, n = 15 −aa −glu, and n = 5 −aa/Refeeding). The experiment was repeated independently three different times. Scale bar: 20 μm. Student's two‐tailed t‐test: ***P‐value < 0.001.
- B
Representative immunoblot using the indicated antibodies from lysates of EndoC‐βH1 cells starved for 1 h of glucose or starved for 1 h and restimulated for 30′ with glucose in the presence or absence of Torin1.
- C
Representative immunoblot for TFEB and TFE3 using lysates from nuclear and cytosolic fractions of INS‐1E cells treated with full medium (FED) or upon amino acid starvation (−aa). PARP1 was used as nuclear fraction loading control, and GAPDH was used as cytosol fraction loading control. Cytosolic fraction (C), Nuclear fraction (N).
- D
Representative immunofluorescence analysis using TFEB antibodies on fixed EndoC‐βH1‐cells treated with full medium (FED) or upon glucose starvation, prior transient transfection with constitutively active Rag GTPase mutants (i.e., HA‐GST‐RagA‐Q66L; HA‐GST‐RagC‐S75L). Transfected cells are indicated with a white arrow. Data are represented as mean ± standard error. Each dot represents a separate field of a representative experiment (n = 5). At least 52 cells were counted in each condition.

- A
Representative immunoblot using lysates from cells infected with control lentivirus (CTRL) of TFEB‐V5‐expressing lentivirus (TFEB‐OE) and analyzed using a V5 antibody. GAPDH was used as a loading control.
- B
TFEB mRNA levels in TFEB‐OE and CTRL EndoC‐βH1 cells incubated with full medium (FED) or starved of amino acids (−aa) for 16 h. Data are represented as mean ± standard error (n = 3–4/group). Student's two‐tailed t‐test: *P‐value < 0.05; **P‐value < 0.01; ***P‐value < 0.001.
- C
Principal component analysis plot of transcriptomic data from TFEB‐OE EndoC‐βH1 cells.
- D
Gene ontology analysis for significantly upregulated (red) and downregulated (green) genes in TFEB‐OE cells compared to control upon amino acid starvation (−aa) for 16 h. Statistically significant hits (FDR < 0.05) are ranked from the most to the least significant.
- E
INS mRNA levels from control (CTRL) and TFEB‐overexpressing (TFEB‐OE) EndoC‐βH1 cells incubated with full medium (FED) or starved of amino acids (−aa) for 16 h. Data are represented as mean ± standard error (n = 3–4/group). Student's two‐tailed t‐test: **P‐value < 0.01; ***P‐value < 0.001.
- F
Representative immunoblot for TFEB and TFE3 in EndoC‐βH1 cells treated with scramble siRNA (siCTRL) or TFEB‐ and TFE3‐targeting siRNA (siTFEB/3). GAPDH was used as a loading control.
- G
TFEB and TFE3 mRNA levels in siTFEB/3 and control EndoC‐βH1 cells incubated with full medium (FED) or starved of amino acids (−aa) for 16 h. Data are represented as mean ± standard error (n = 4/group). Student's two‐tailed t‐test: **P‐value < 0.01; ***P‐value < 0.001.
- H
Principal component analysis plot of transcriptomic data from siTFEB/TFE3 EndoC‐βH1 cells.
- I
Gene ontology analysis for significantly upregulated (red) and downregulated (green) genes in cells depleted for TFEB and TFE3 compared to control upon amino acid starvation (−aa) for 16 h.
- J
INS mRNA levels from EndoC‐βH1 cells treated with scramble siRNA (siCTRL) or TFEB and TFE3‐targeting siRNA (siTFEB/3) incubated with full medium (FED) or starved of amino acids (−aa) for 16 h. Data are represented as mean ± standard error (n = 4/group). Student's two‐tailed t‐test: *P‐value < 0.05.
- K
Venn diagram showing the comparison of the datasets of TFEB‐OE versus TFEB/3‐depleted EndoC‐βH1 cells.
- L
Heatmap showing the 751 DEGs significantly regulated in opposite correlation in TFEB‐OE versus TFEB/3‐depleted EndoC‐βH1 cells. The lane corresponding to the insulin gene (INS) is indicated.
- M
Gene ontology analysis for significantly downregulated genes in siTFEB/3 and upregulated in TFEB‐OE cells in opposite correlation.

- A
Representative immunoblot of lysates from INS‐1E cells infected with hTFEB‐Flag‐expressing lentivirus (TFEB‐OE) or control (CTRL) cells.
- B
Principal component analysis plot of transcriptomic data from TFEB‐OE INS1 cells.
- C
Gene ontology analysis for significantly upregulated (red) and downregulated (green) genes in TFEB‐overexpressing (TFEB‐OE) cells compared to control (CTRL) upon amino acid starvation (−aa) for 16 h.
- D
INS1 and INS2 mRNA levels from control (CTRL) or TFEB‐overexpressing (TFEB‐OE) INS1 cells incubated with full medium (FED) or upon amino acid starvation (−aa) for 16 h. Each dot represents one mouse (n = 3‐4/group). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- E
Representative immunoblot for TFEB and TFE3 in DKO cells. GAPDH was used as a loading control.
- F
Principal component analysis plot of transcriptomic data from TFEB/TFE3 DKO INS1 cells.
- G
Gene enrichment analysis for significantly upregulated (green) and downregulated (red) genes from RNA seq data of DKO cells in comparison to control cells upon amino acid starvation (−aa) for 16 h.
- H
INS1 and INS2 mRNA levels from CTRL or TFEB/TFE3‐DKO INS1 cells incubated with full medium (FED) or upon amino acid starvation (−aa) for 16 h. Each dot represents one mouse (n = 3‐4/group). Data are represented as mean ± standard error Student's two‐tailed t‐test: *P‐value < 0.05; ***P‐value < 0.001.
- I
Venn diagram showing the comparison of the datasets of TFEB‐overexpressing (TFEB‐OE) versus TFEB/3 DKO INS1 cells.
- J
Heatmap showing the 550 DEGs significantly regulated in opposite correlation in TFEB‐OE versus TFEB/3 DKO INS1 cells. The lanes corresponding to insulin genes (Ins1 and Ins2) are indicated.

- A
Graphic representation of the different genomic sites of TFEB binding in EndoC‐βH1 cells upon 16 h amino acid starvation.
- B
Venn diagram of genes upregulated upon TFEB overexpression and downregulated in siTFEB/3 and TFEB‐binding sites located inside proximal promoters (−1,000 + 300).
- C
Gene ontology analysis for TFEB promoter‐bound direct targets.
- D, E
Alignment of the TFEB ChIP‐seq track, aligned with ChIP‐seq tracks of other known INS regulators (MAFB, NKX2.2, NKX6.1, and PDX1), at the INS locus. Super‐enhancer, black box.

- A
Top genes whose promoters are bound by TFEB upon fasting and upregulated upon TFEB upregulation in EndoC‐BH1 cells sorted by gene categories. Genes in red are previously identified TFEB direct targets.
- B–F
On the left, alignment of the TFEB ChIP‐seq track at the indicated gene loci. On the right, mRNA levels of the indicated genes from EndoC‐βH1 cells treated with scramble siRNA (siCTRL) or TFEB‐ and TFE3‐targeting siRNA (siTFEB/3) or TFEB‐OE compared to control EndoC‐βH1 cells incubated with full medium (FED) or upon amino acid starva tion (−aa) for 16 h. Data are represented as mean ± standard error. Each dot represents an independent experiment, ***P < 0.001; Student's two‐tailed unpaired t‐test.
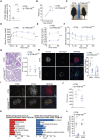
- A
mRNA levels of TFEB in isolated islets from control and βTFEBOELOW mice. Each dot represents one mouse (n = 3–4/group). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- B
Weight gain of control (n = 20) and βTFEBOELOW mice (n = 14). Data are represented as mean ± standard error. Two‐way ANOVA: ***P‐value < 0.001.
- C
Representative images of control and βTFEBOELOW mice showing different body sizes.
- D
Glucose tolerance test (GTT) of control (n = 15) and βTFEBOELOW (n = 11) mice. Data are represented as mean ± standard error. Two‐way ANOVA: ***P‐value < 0.001.
- E
Glucose‐stimulated insulin secretion (GSIS) for control (n = 9) and βTFEBOELOW (n = 7) mice. Data are represented as mean ± standard error. Two‐way ANOVA: ***P‐value < 0.001.
- F
Insulin tolerance test (ITT) normalized to T0 in control and βTFEBOELOW mice (n = 9/group). Data are represented as mean ± standard error. Two‐way ANOVA: *P‐value < 0.05.
- G
Representative images of pancreas slides from control and βTFEBOELOW mice stained with hematoxylin/eosin and quantification of islet area (n = 6/group; n = 5–10 islets per mouse). Data are represented as mean ± standard error.
- H
Representative immunofluorescence picture using insulin and glucagon antibodies on fixed pancreas of control and βTFEBOELOW mice (scale bar 50 μm). The graph shows the quantification of the relative insulin (INS+)‐ and glucagon (GCG+)‐positive area from islets of control and βTFEBOELOW mice. Each dot represents an islet (n = 15 for CTRL and n = 12 for βTFEBOELOW). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- I, J
Representative immunofluorescence images for TFEB and UCN3 (I) on fixed pancreas of control and βTFEBOELOW mice (scale bar 20 μm) and quantification of the UCN3+ area per islet in control and βTFEBOELOW pancreas (n = 3/group; n = 3–14 islets per mouse) (J). Data are represented as mean ± standard error. Student's two‐tailed t‐test: *P‐value < 0.05.
- K
Gene ontology analysis of RNA‐seq data of isolated islets from control and βTFEBOELOW mice.
- L
mRNA levels of indicated genes assessed on isolated islets from control and βTFEBOELOW mice. Each dot represents one mouse (n = 3–4/group). Data are represented as mean ± standard error. Student's two‐tailed t‐test: **P‐value < 0.01.

- A
mRNA levels of TFEB in isolated islets from control and βTFEBOEHIGH mice. Each dot represents a mouse (n = 4‐5/group). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- B
Weight of control and βTFEBOEHIGH mice. Each dot represents a mouse (n = 9 for CTRL and n = 5 for βTFEBOEHIGH mice). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- C
Representative images of control and βTFEBOEHIGH mice showing different body sizes.
- D
Glucose tolerance test (GTT) of control (n = 12) and βTFEBOEHIGH (n = 7) mice. Data are represented as mean ± standard error. Two‐way ANOVA: ***P‐value < 0.001.
- E
Glucose‐stimulated insulin secretion (GSIS) for control (n = 11) and βTFEBOEHIGH (n = 7) mice. Data are represented as mean ± standard error. Two‐way ANOVA: ***P‐value < 0.001.
- F
mRNA levels of Ins1 and Ins2 in isolated islets from control and βTFEBOEHIGH mice. Each dot represents a mouse (n = 5/group). Data are represented as mean ± standard error. Student's two‐tailed t‐test: ****P‐value < 0.0001.
- G
Representative images of pancreas slides from control and βTFEBOEHIGH mice, stained with hematoxylin/eosin and relative quantification of islet area (n = 3‐4/group). Data are represented as mean ± standard error.

- A
Representative images of RNA scope analysis for Ins2 mRNA in pancreas from control and β‐DKO mice in fed and starved conditions with relative quantification (n = 3/group). Each dot represents one mouse. Data are represented as mean ± standard error. Student's two‐tailed t‐test: *P‐value < 0.05; ***P‐value < 0.001.
- B
Representative images of pancreas slides from control and β‐DKO mice stained with hematoxylin/eosin and quantification of islet area (n = 6/group; n = 11–25 islets per mouse). Each dot represents one mouse.
- C
Representative immunofluorescence images for UCN3 and quantification of the UCN3+ area per islet in control and β β‐DKO pancreas (n = 3/group; n = 8–25 islets per mouse). Each dot represents one mouse. Data are represented as mean ± standard error. Student's two‐tailed t‐test: ***P‐value < 0.001.
- D
Body weight of control and β‐DKO mice at 8 weeks of age.
- E
Glucose tolerance test (GTT) of control and β‐DKO mice. Each dot represents one mouse (n = 8/group). Two‐way ANOVA: ***P‐value < 0.001.
References
-
- Alvarez‐Dominguez JR, Donaghey J, Rasouli N, Kenty JHR, Helman A, Charlton J, Straubhaar JR, Meissner A, Melton DA (2020) Circadian entrainment triggers maturation of human in vitro islets. Cell Stem Cell 26: 108–122.e10 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

