The Drosophila homolog of APP promotes Dscam expression to drive axon terminal growth, revealing interaction between Down syndrome genes
- PMID: 37712356
- PMCID: PMC10508694
- DOI: 10.1242/dmm.049725
The Drosophila homolog of APP promotes Dscam expression to drive axon terminal growth, revealing interaction between Down syndrome genes
Abstract
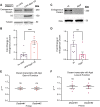
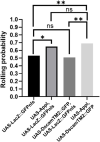
Down syndrome (DS) is caused by triplication of human chromosome 21 (HSA21). Although several HSA21 genes have been found to be responsible for aspects of DS, whether and how HSA21 genes interact with each other is poorly understood. DS patients and animal models present with a number of neurological changes, including aberrant connectivity and neuronal morphology. Previous studies have indicated that amyloid precursor protein (APP) and Down syndrome cell adhesion molecule (DSCAM) regulate neuronal morphology and contribute to neuronal aberrations in DS. Here, we report the functional interaction between the Drosophila homologs of these two genes, Amyloid precursor protein-like (Appl) and Dscam (Dscam1). We show that Appl requires Dscam to promote axon terminal growth in sensory neurons. Moreover, Appl increases Dscam protein expression post-transcriptionally. We further demonstrate that regulation of Dscam by Appl does not require the Appl intracellular domain or second extracellular domain. This study presents an example of functional interactions between HSA21 genes, providing insights into the pathogenesis of neuronal aberrations in DS.
Keywords: Drosophila; APP; Amyloid precursor protein; Axon; Down syndrome; Down syndrome cell adhesion molecule; Dscam.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
-
- Canfield, M. A., Honein, M. A., Yuskiv, N., Xing, J., Mai, C. T., Collins, J. S., Devine, O., Petrini, J., Ramadhani, T. A., Hobbs, C. A.et al. (2006). National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999-2001. Birth Defects Res. A Clin. Mol. Teratol. 76, 747-756. 10.1002/bdra.20294 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

