Precision detection of recent HIV infections using high-throughput genomic incidence assay
- PMID: 37712639
- PMCID: PMC10580985
- DOI: 10.1128/spectrum.02285-23
Precision detection of recent HIV infections using high-throughput genomic incidence assay
Abstract
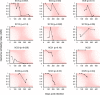
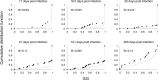
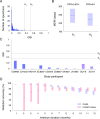
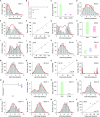
HIV incidence is a key measure for tracking disease spread and identifying populations and geographic regions where new infections are most concentrated. The HIV sequence population provides a robust signal for the stage of infection. Large-scale and high-precision HIV sequencing is crucial for effective genomic incidence surveillance. We produced 1,034 full-length envelope gene sequences from a seroconversion cohort by conducting HIV microdrop sequencing and measuring the genomic incidence assay's genome similarity index (GSI) dynamics. The measured dynamics of 9 of 12 individuals aligned with the GSI distribution estimated independently using 417 publicly available incident samples. We enhanced the capacity to identify individuals with recent infections, achieving predicted detection accuracies of 92% (89%-94%) for cases within 6 months and 81% (74%-87%) for cases within 9 months. These accuracy levels agreed with the observed detection accuracy intervals of an independent validation data set. Additionally, we produced 131 full-length envelope gene sequences from eight individuals with chronic HIV infection. This analysis confirmed a false recency rate (FRR) of 0%, which was consistent with 162 publicly available chronic samples. The mean duration of recent infection (MDRI) was 238 (209-267) days, indicating an 83% improvement in performance compared to current recent infection testing algorithms. The shifted Poisson mixture model was then used to estimate the time since infection, and the model estimates showed an 88% consistency with the days post infection derived from HIV RNA test dates and/or seroconversion dates. HIV microdrop sequencing provides unique prospects for large-scale incidence surveillance using high-throughput sequencing. IMPORTANCE Accurate identification of recently infected individuals is vital for prioritizing specific populations for interventions, reducing onward transmission risks, and optimizing public health services. However, current HIV-specific antibody-based methods have not been satisfactory in accurately identifying incident cases, hindering the use of HIV recency testing for prevention efforts and partner protection. Genomic incidence assays offer a promising alternative for identifying recent infections. In our study, we used microdroplet technologies to produce a large number of complete HIV envelope gene sequences, enabling the accurate detection of early infection signs. We assessed the dynamics of the incidence assay's metrics and compared them with statistical models. Our approach demonstrated high accuracy in identifying individuals with recent infections, achieving predicted detection rates exceeding 90% within 6 months and over 80% within 9 months of infection. This high-resolution method holds significant potential for enhancing the effectiveness of HIV incidence screening for case-based surveillance in public health initiatives.
Keywords: HIV incidence; genomic epidemiology; genomic surveillance; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Busch MP, Pilcher CD, Mastro TD, Kaldor J, Vercauteren G, Rodriguez W, Rousseau C, Rehle TM, Welte A, Averill MD, Garcia Calleja JM, WHO Working Group on HIV Incidence Assays . 2010. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS 24:2763–2771. doi: 10.1097/QAD.0b013e32833f1142 - DOI - PubMed
-
- Grabowski MK, Serwadda DM, Gray RH, Nakigozi G, Kigozi G, Kagaayi J, Ssekubugu R, Nalugoda F, Lessler J, Lutalo T, Galiwango RM, Makumbi F, Kong X, Kabatesi D, Alamo ST, Wiersma S, Sewankambo NK, Tobian AAR, Laeyendecker O, Quinn TC, Reynolds SJ, Wawer MJ, Chang LW, Rakai Health Sciences Program . 2017. HIV prevention efforts and incidence of HIV in Uganda. N Engl J Med 377:2154–2166. doi: 10.1056/NEJMoa1702150 - DOI - PMC - PubMed
-
- Wirtz AL, Humes E, Althoff KN, Poteat TC, Radix A, Mayer KH, Schneider JS, Haw JS, Wawrzyniak AJ, Cannon CM, Stevenson M, Cooney EE, Adams D, Case J, Beyrer C, Laeyendecker O, Rodriguez AE, Reisner SL, American Cohort to Study HIV Acquisition Among Transgender Women (LITE) Study Group . 2023. HIV incidence and mortality in transgender women in the eastern and Southern USA: a multisite cohort study. Lancet HIV 10:e308–e319. doi: 10.1016/S2352-3018(23)00008-5 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

