Control of Helicobacter pylori with engineered probiotics secreting selective guided antimicrobial peptides
- PMID: 37712669
- PMCID: PMC10580918
- DOI: 10.1128/spectrum.02014-23
Control of Helicobacter pylori with engineered probiotics secreting selective guided antimicrobial peptides
Abstract
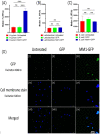
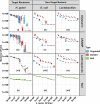
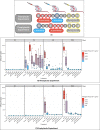
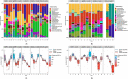
Helicobacter pylori is the primary cause of 78% of gastric cancer cases, providing an opportunity to prevent cancer by controlling a single bacterial pathogen within the complex gastric microbiota. We developed highly selective antimicrobial agents against H. pylori by fusing an H. pylori-binding guide peptide (MM1) to broad-spectrum antimicrobial peptides. The common dairy probiotic Lactococcus lactis was then engineered to secrete these guided antimicrobial peptides (gAMPs). When co-cultured in vitro with H. pylori, the gAMP probiotics lost no toxicity compared to unguided AMP probiotics against the target, H. pylori, while losing >90% of their toxicity against two tested off-target bacteria. To test binding to H. pylori, the MM1 guide was fused to green fluorescent protein (GFP), resulting in enhanced binding compared to unguided GFP as measured by flow cytometry. In contrast, MM1-GFP showed no increased binding over GFP against five different off-target bacteria. These highly selective gAMP probiotics were then tested by oral gavage in mice infected with H. pylori. As a therapy, the probiotics outperformed antibiotic treatment, effectively eliminating H. pylori in just 5 days, and also protected mice from challenge infection as a prophylactic. As expected, the gAMP probiotics were as toxic against H. pylori as the unguided AMP probiotics. However, a strong rebound in gastric species diversity was found with both the selective gAMP probiotics and the non-selective AMP probiotics. Eliminating the extreme microbial dysbiosis caused by H. pylori appeared to be the major factor in diversity recovery. IMPORTANCE Alternatives to antibiotics in the control of Helicobacter pylori and the prevention of gastric cancer are needed. The high prevalence of H. pylori in the human population, the induction of microbial dysbiosis by antibiotics, and increasing antibiotic resistance call for a more sustainable approach. By selectively eliminating the pathogen and retaining the commensal community, H. pylori control may be achieved without adverse health outcomes. Antibiotics are typically used as a therapeutic post-infection, but a more targeted, less disruptive approach could be used as a long-term prophylactic against H. pylori or, by extension, against other gastrointestinal pathogens. Furthermore, the modular nature of the guided antimicrobial peptide (gAMP) technology allows for the substitution of different guides for different pathogens and the use of a cocktail of gAMPs to avoid the development of pathogen resistance.
Keywords: Helicobacter pylori; antimicrobial; dysbiosis; microbiome; mouse; probiotic; selective.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- International agency for research on cancer . 2012. A review of human carcinogens - PubMed
-
- Yang L, Kartsonaki C, Yao P, de Martel C, Plummer M, Chapman D, Guo Y, Clark S, Walters RG, Chen Y, Pei P, Lv J, Yu C, Jeske R, Waterboer T, Clifford GM, Franceschi S, Peto R, Hill M, Li L, Millwood IY, Chen Z, China Kadoorie Biobank Collaborative Group . 2021. The relative and attributable risks of cardia and non-cardia gastric cancer associated with Helicobacter pylori infection in China: a case-cohort study. Lancet Public Health 6:e888–e896. doi: 10.1016/S2468-2667(21)00164-X - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

