PA2G4/EBP1 ubiquitination by PRKN/PARKIN promotes mitophagy protecting neuron death in cerebral ischemia
- PMID: 37712850
- PMCID: PMC10813645
- DOI: 10.1080/15548627.2023.2259215
PA2G4/EBP1 ubiquitination by PRKN/PARKIN promotes mitophagy protecting neuron death in cerebral ischemia
Abstract
Cerebral ischemia induces massive mitochondrial damage, leading to neuronal death. The elimination of damaged mitochondria via mitophagy is critical for neuroprotection. Here we show that the level of PA2G4/EBP1 (proliferation-associated 2G4) was notably increased early during transient middle cerebral artery occlusion and prevented neuronal death by eliciting cerebral ischemia-reperfusion (IR)-induced mitophagy. Neuron-specific knockout of Pa2g4 increased infarct volume and aggravated neuron loss with impaired mitophagy and was rescued by introduction of adeno-associated virus serotype 2 expressing PA2G4/EBP1. We determined that PA2G4/EBP1 is ubiquitinated on lysine 376 by PRKN/PARKIN on the damaged mitochondria and interacts with receptor protein SQSTM1/p62 for mitophagy induction. Thus, our study suggests that PA2G4/EBP1 ubiquitination following cerebral IR-injury promotes mitophagy induction, which may be implicated in neuroprotection.Abbreviations: AAV: adeno-associated virus; ACTB: actin beta; BNIP3L/NIX: BCL2 interacting protein 3 like; CA1: Cornu Ammonis 1; CASP3: caspase 3; CCCP: carbonyl cyanide m-chlorophenyl hydrazone; DMSO: dimethyl sulfoxide; PA2G4/EBP1: proliferation-associated 2G4; FUNDC1: FUN14 domain containing 1; IB: immunoblotting; ICC: immunocytochemistry; IHC: immunohistochemistry; IP: immunoprecipitation; MCAO: middle cerebral artery occlusion; MEF: mouse embryonic fibroblast; OGD: oxygen-glucose deprivation; PRKN/PARKIN: parkin RBR E3 ubiquitin protein ligase; PINK1: PTEN induced kinase 1; RBFOX3/NeuN: RNA binding fox-1 homolog 3; SQSTM1/p62: sequestosome 1; TIMM23: translocase of inner mitochondrial membrane 23; TOMM20: translocase of outer mitochondrial membrane 20; TUBB: tubulin beta class I; WT: wild-type.
Keywords: Ischemia; PA2G4/EBP1; PRKN/PARKIN; SQSTM1/p62; mitophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
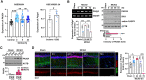
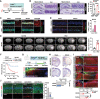

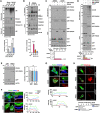


References
-
- Kirino T, Sano K.. Fine structural nature of delayed neuronal death following ischemia in the gerbil hippocampus. Acta Neuropathol. 1984;62:209–18. - PubMed
-
- Tanaka K, Ludwig LM, Krolikowski JG, et al. Isoflurane produces delayed preconditioning against myocardial ischemia and reperfusion injury: role of cyclooxygenase-2. Anesthesiology. 2004;100:525–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
