BATF relieves hepatic steatosis by inhibiting PD1 and promoting energy metabolism
- PMID: 37712938
- PMCID: PMC10503959
- DOI: 10.7554/eLife.88521
BATF relieves hepatic steatosis by inhibiting PD1 and promoting energy metabolism
Abstract
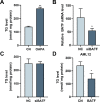
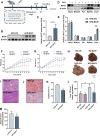
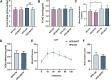
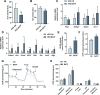
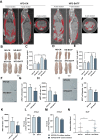
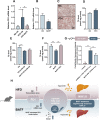
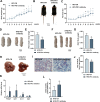
The rising prevalence of nonalcoholic fatty liver disease (NAFLD) has become a global health threat that needs to be addressed urgently. Basic leucine zipper ATF-like transcription factor (BATF) is commonly thought to be involved in immunity, but its effect on lipid metabolism is not clear. Here, we investigated the function of BATF in hepatic lipid metabolism. BATF alleviated high-fat diet (HFD)-induced hepatic steatosis and inhibited elevated programmed cell death protein (PD)1 expression induced by HFD. A mechanistic study confirmed that BATF regulated fat accumulation by inhibiting PD1 expression and promoting energy metabolism. PD1 antibodies alleviated hepatic lipid deposition. In conclusion, we identified the regulatory role of BATF in hepatic lipid metabolism and that PD1 is a target for alleviation of NAFLD. This study provides new insights into the relationship between BATF, PD1, and NAFLD.
Keywords: BATF; NAFLD; PD1; cell biology; immune; mouse.
© 2023, Zhang, Liao et al.
Conflict of interest statement
ZZ, QL, TP, LY, ZL, SS, SL, MH, YL, TD, YL, LZ No competing interests declared
Figures


Update of
- doi: 10.1101/2023.04.18.537352
- doi: 10.7554/eLife.88521.1
- doi: 10.7554/eLife.88521.2
References
-
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. The Journal of Experimental Medicine. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

