Structure/function studies of the NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding
- PMID: 37712998
- PMCID: PMC10504179
- DOI: 10.1007/s00792-023-01309-z
Structure/function studies of the NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding
Abstract
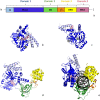
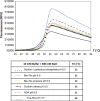
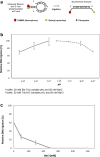
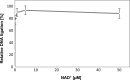
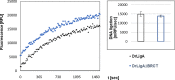
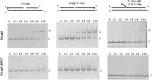
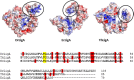
Bacterial NAD+-dependent DNA ligases (LigAs) are enzymes involved in replication, recombination, and DNA-repair processes by catalyzing the formation of phosphodiester bonds in the backbone of DNA. These multidomain proteins exhibit four modular domains, that are highly conserved across species, with the BRCT (breast cancer type 1 C-terminus) domain on the C-terminus of the enzyme. In this study, we expressed and purified both recombinant full-length and a C-terminally truncated LigA from Deinococcus radiodurans (DrLigA and DrLigA∆BRCT) and characterized them using biochemical and X-ray crystallography techniques. Using seeds of DrLigA spherulites, we obtained ≤ 100 µm plate crystals of DrLigA∆BRCT. The crystal structure of the truncated protein was obtained at 3.4 Å resolution, revealing DrLigA∆BRCT in a non-adenylated state. Using molecular beacon-based activity assays, we demonstrated that DNA ligation via nick sealing remains unaffected in the truncated DrLigA∆BRCT. However, DNA-binding assays revealed a reduction in the affinity of DrLigA∆BRCT for dsDNA. Thus, we conclude that the flexible BRCT domain, while not critical for DNA nick-joining, plays a role in the DNA binding process, which may be a conserved function of the BRCT domain in LigA-type DNA ligases.
Keywords: BRCT; DNA ligase A; DNA nick-joining; Protein–DNA binding; X-ray crystallography.
© 2023. The Author(s).
Figures


Similar articles
-
Enzymes involved in DNA ligation and end-healing in the radioresistant bacterium Deinococcus radiodurans.BMC Mol Biol. 2007 Aug 16;8:69. doi: 10.1186/1471-2199-8-69. BMC Mol Biol. 2007. PMID: 17705817 Free PMC article.
-
Biochemical characterization of two DNA ligases from Deinococcus radiodurans.Protein Pept Lett. 2008;15(6):600-5. doi: 10.2174/092986608784967010. Protein Pept Lett. 2008. PMID: 18680457
-
1H, 15N and 13C resonance backbone and side-chain assignments and secondary structure determination of the BRCT domain of Mtb LigA.Biomol NMR Assign. 2024 Jun;18(1):105-109. doi: 10.1007/s12104-024-10175-5. Epub 2024 Apr 30. Biomol NMR Assign. 2024. PMID: 38689205
-
ATP-dependent DNA ligases.Genome Biol. 2002;3(4):REVIEWS3005. doi: 10.1186/gb-2002-3-4-reviews3005. Epub 2002 Mar 19. Genome Biol. 2002. PMID: 11983065 Free PMC article. Review.
-
DNA ligases in the repair and replication of DNA.Mutat Res. 2000 Aug 30;460(3-4):301-18. doi: 10.1016/s0921-8777(00)00033-1. Mutat Res. 2000. PMID: 10946235 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

