Laboratory evolution, transcriptomics, and modeling reveal mechanisms of paraquat tolerance
- PMID: 37713311
- PMCID: PMC10591938
- DOI: 10.1016/j.celrep.2023.113105
Laboratory evolution, transcriptomics, and modeling reveal mechanisms of paraquat tolerance
Abstract
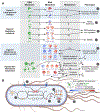
Relationships between the genome, transcriptome, and metabolome underlie all evolved phenotypes. However, it has proved difficult to elucidate these relationships because of the high number of variables measured. A recently developed data analytic method for characterizing the transcriptome can simplify interpretation by grouping genes into independently modulated sets (iModulons). Here, we demonstrate how iModulons reveal deep understanding of the effects of causal mutations and metabolic rewiring. We use adaptive laboratory evolution to generate E. coli strains that tolerate high levels of the redox cycling compound paraquat, which produces reactive oxygen species (ROS). We combine resequencing, iModulons, and metabolic models to elucidate six interacting stress-tolerance mechanisms: (1) modification of transport, (2) activation of ROS stress responses, (3) use of ROS-sensitive iron regulation, (4) motility, (5) broad transcriptional reallocation toward growth, and (6) metabolic rewiring to decrease NADH production. This work thus demonstrates the power of iModulon knowledge mapping for evolution analysis.
Keywords: CP: Microbiology; adaptive laboratory evolution; big data analytics; computational biology; oxidative stress; paraquat; systems biology; transcriptional regulatory networks; transcriptomics.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



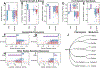


Similar articles
-
Modulating bacterial function utilizing A knowledge base of transcriptional regulatory modules.Nucleic Acids Res. 2024 Oct 14;52(18):11362-11377. doi: 10.1093/nar/gkae742. Nucleic Acids Res. 2024. PMID: 39193902 Free PMC article.
-
Systems biology of competency in Vibrio natriegens is revealed by applying novel data analytics to the transcriptome.Cell Rep. 2023 Jun 27;42(6):112619. doi: 10.1016/j.celrep.2023.112619. Epub 2023 Jun 6. Cell Rep. 2023. PMID: 37285268
-
Meta-analysis Driven Strain Design for Mitigating Oxidative Stresses Important in Biomanufacturing.ACS Synth Biol. 2024 Jul 19;13(7):2045-2059. doi: 10.1021/acssynbio.3c00572. Epub 2024 Jun 27. ACS Synth Biol. 2024. PMID: 38934464 Free PMC article.
-
Diagnosis and mitigation of the systemic impact of genome reduction in Escherichia coli DGF-298.mBio. 2024 Oct 16;15(10):e0087324. doi: 10.1128/mbio.00873-24. Epub 2024 Aug 29. mBio. 2024. PMID: 39207109 Free PMC article.
-
Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate.J Bacteriol. 2001 Jul;183(13):3890-902. doi: 10.1128/JB.183.13.3890-3902.2001. J Bacteriol. 2001. PMID: 11395452 Free PMC article.
Cited by
-
Trade-off between resistance and persistence in high cell density cultures.mSystems. 2025 Jul 22;10(7):e0032325. doi: 10.1128/msystems.00323-25. Epub 2025 Jun 13. mSystems. 2025. PMID: 40511936 Free PMC article.
-
The hallmarks of a tradeoff in transcriptomes that balances stress and growth functions.mSystems. 2024 Jul 23;9(7):e0030524. doi: 10.1128/msystems.00305-24. Epub 2024 Jun 3. mSystems. 2024. PMID: 38829048 Free PMC article.
-
Revisiting oxygen toxicity: evolution and adaptation to superoxide in a SOD-deficient bacterial pathogen.mBio. 2025 Aug 13;16(8):e0064525. doi: 10.1128/mbio.00645-25. Epub 2025 Jul 23. mBio. 2025. PMID: 40698983 Free PMC article.
-
Repeated glucose oscillations in high cell-density cultures influence stress-related functions of Escherichia coli.PNAS Nexus. 2024 Sep 2;3(9):pgae376. doi: 10.1093/pnasnexus/pgae376. eCollection 2024 Sep. PNAS Nexus. 2024. PMID: 39285935 Free PMC article.
-
Modulating bacterial function utilizing A knowledge base of transcriptional regulatory modules.Nucleic Acids Res. 2024 Oct 14;52(18):11362-11377. doi: 10.1093/nar/gkae742. Nucleic Acids Res. 2024. PMID: 39193902 Free PMC article.
References
Additional resources
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

