AFsample: improving multimer prediction with AlphaFold using massive sampling
- PMID: 37713472
- PMCID: PMC10534052
- DOI: 10.1093/bioinformatics/btad573
AFsample: improving multimer prediction with AlphaFold using massive sampling
Abstract
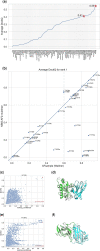
Summary: The AlphaFold2 neural network model has revolutionized structural biology with unprecedented performance. We demonstrate that by stochastically perturbing the neural network by enabling dropout at inference combined with massive sampling, it is possible to improve the quality of the generated models. We generated ∼6000 models per target compared with 25 default for AlphaFold-Multimer, with v1 and v2 multimer network models, with and without templates, and increased the number of recycles within the network. The method was benchmarked in CASP15, and compared with AlphaFold-Multimer v2 it improved the average DockQ from 0.41 to 0.55 using identical input and was ranked at the very top in the protein assembly category when compared with all other groups participating in CASP15. The simplicity of the method should facilitate the adaptation by the field, and the method should be useful for anyone interested in modeling multimeric structures, alternate conformations, or flexible structures.
Availability and implementation: AFsample is available online at http://wallnerlab.org/AFsample.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
References
-
- Cramer P. AlphaFold2 and the future of structural biology. Nat Struct Mol Biol 2021;28:704–5. - PubMed
-
- Gal Y, Ghahramani Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. PMLR 2016;48:1050–1059.

