A multi-scale expression and regulation knowledge base for Escherichia coli
- PMID: 37713610
- PMCID: PMC10602906
- DOI: 10.1093/nar/gkad750
A multi-scale expression and regulation knowledge base for Escherichia coli
Abstract
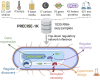
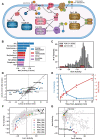
Transcriptomic data is accumulating rapidly; thus, scalable methods for extracting knowledge from this data are critical. Here, we assembled a top-down expression and regulation knowledge base for Escherichia coli. The expression component is a 1035-sample, high-quality RNA-seq compendium consisting of data generated in our lab using a single experimental protocol. The compendium contains diverse growth conditions, including: 9 media; 39 supplements, including antibiotics; 42 heterologous proteins; and 76 gene knockouts. Using this resource, we elucidated global expression patterns. We used machine learning to extract 201 modules that account for 86% of known regulatory interactions, creating the regulatory component. With these modules, we identified two novel regulons and quantified systems-level regulatory responses. We also integrated 1675 curated, publicly-available transcriptomes into the resource. We demonstrated workflows for analyzing new data against this knowledge base via deconstruction of regulation during aerobic transition. This resource illuminates the E. coli transcriptome at scale and provides a blueprint for top-down transcriptomic analysis of non-model organisms.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
The Escherichia coli transcriptome mostly consists of independently regulated modules.Nat Commun. 2019 Dec 4;10(1):5536. doi: 10.1038/s41467-019-13483-w. Nat Commun. 2019. PMID: 31797920 Free PMC article.
-
iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning.Nucleic Acids Res. 2021 Jan 8;49(D1):D112-D120. doi: 10.1093/nar/gkaa810. Nucleic Acids Res. 2021. PMID: 33045728 Free PMC article.
-
Genomewide Stabilization of mRNA during a "Feast-to-Famine" Growth Transition in Escherichia coli.mSphere. 2020 May 20;5(3):e00276-20. doi: 10.1128/mSphere.00276-20. mSphere. 2020. PMID: 32434841 Free PMC article.
-
Escherichia coli--a model system that benefits from and contributes to the evolution of proteomics.Biotechnol Bioeng. 2003 Dec 30;84(7):801-14. doi: 10.1002/bit.10848. Biotechnol Bioeng. 2003. PMID: 14708121 Review.
-
Extending knowledge of Escherichia coli metabolism by modeling and experiment.Genome Biol. 2003;4(11):235. doi: 10.1186/gb-2003-4-11-235. Epub 2003 Oct 28. Genome Biol. 2003. PMID: 14611652 Free PMC article. Review.
Cited by
-
Machine learning uncovers the Pseudomonas syringae transcriptome in microbial communities and during infection.mSystems. 2023 Oct 26;8(5):e0043723. doi: 10.1128/msystems.00437-23. Epub 2023 Aug 28. mSystems. 2023. PMID: 37638727 Free PMC article.
-
A Causal Regulation Modeling Algorithm for Temporal Events with Application to Escherichia coli's Aerobic to Anaerobic Transition.Int J Mol Sci. 2024 May 22;25(11):5654. doi: 10.3390/ijms25115654. Int J Mol Sci. 2024. PMID: 38891842 Free PMC article.
-
PGBTR: a powerful and general method for inferring bacterial transcriptional regulatory networks.BMC Genomics. 2025 Aug 1;26(1):712. doi: 10.1186/s12864-025-11863-9. BMC Genomics. 2025. PMID: 40750847 Free PMC article.
-
Deciphering the proteome of Escherichia coli K-12: Integrating transcriptomics and machine learning to annotate hypothetical proteins.Comput Struct Biotechnol J. 2025 Jul 24;27:3565-3578. doi: 10.1016/j.csbj.2025.07.036. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40821719 Free PMC article.
-
The Environment-Dependent Regulatory Landscape of the E. coli Genome.ArXiv [Preprint]. 2025 May 13:arXiv:2505.08764v1. ArXiv. 2025. PMID: 40463697 Free PMC article. Preprint.

