Characterization of novel recombinant mycobacteriophages derived from homologous recombination between two temperate phages
- PMID: 37713616
- PMCID: PMC10700106
- DOI: 10.1093/g3journal/jkad210
Characterization of novel recombinant mycobacteriophages derived from homologous recombination between two temperate phages
Abstract
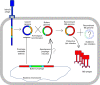
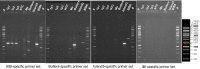
Comparative analyses of mycobacteriophage genomes reveals extensive genetic diversity in genome organization and gene content, contributing to widespread mosaicism. We previously reported that the prophage of mycobacteriophage Butters (cluster N) provides defense against infection by Island3 (subcluster I1). To explore the anti-Island3 defense mechanism, we attempted to isolate Island3 defense escape mutants on a Butters lysogen, but only uncovered phages with recombinant genomes comprised of regions of Butters and Island3 arranged from left arm to right arm as Butters-Island3-Butters (BIBs). Recombination occurs within two distinct homologous regions that encompass lysin A, lysin B, and holin genes in one segment, and RecE and RecT genes in the other. Structural genes of mosaic BIB genomes are contributed by Butters while the immunity cassette is derived from Island3. Consequently, BIBs are morphologically identical to Butters (as shown by transmission electron microscopy) but are homoimmune with Island3. Recombinant phages overcome antiphage defense and silencing of the lytic cycle. We leverage this observation to propose a stratagem to generate novel phages for potential therapeutic use.
Keywords: bacteriophage evolution; homologous recombination; mycobacteriophage genomics; phage genetic diversity; phage genome mosaicism; phage therapy.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures




References
-
- https://phagesdb.org/ The Actinobacteriophage Database | Home.
-
- Alcoser TA, Alexander LM, Bayles IM, Campbell IW, Cohen LB, English BV, Filliger LZ, Fox TM, Guerra SL, He S, et al. . 2010. Mycobacterium phage Island3, complete genome. GenBank: HM152765.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
