OTUD5 limits replication fork instability by organizing chromatin remodelers
- PMID: 37713620
- PMCID: PMC10602872
- DOI: 10.1093/nar/gkad732
OTUD5 limits replication fork instability by organizing chromatin remodelers
Abstract
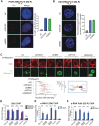
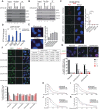
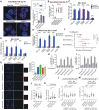
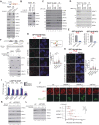
Proper regulation of replication fork progression is important for genomic maintenance. Subverting the transcription-induced conflicts is crucial in preserving the integrity of replication forks. Various chromatin remodelers, such as histone chaperone and histone deacetylases are known to modulate replication stress, but how these factors are organized or collaborate are not well understood. Here we found a new role of the OTUD5 deubiquitinase in limiting replication stress. We found that OTUD5 is recruited to replication forks, and its depletion causes replication fork stress. Through its C-terminal disordered tail, OTUD5 assembles a complex containing FACT, HDAC1 and HDAC2 at replication forks. A cell line engineered to specifically uncouple FACT interaction with OTUD5 exhibits increases in FACT loading onto chromatin, R-loop formation, and replication fork stress. OTUD5 mediates these processes by recruiting and stabilizing HDAC1 and HDAC2, which decreases H4K16 acetylation and FACT recruitment. Finally, proteomic analysis revealed that the cells with deficient OTUD5-FACT interaction activates the Fanconi Anemia pathway for survival. Altogether, this study identified a new interaction network among OTUD5-FACT-HDAC1/2 that limits transcription-induced replication stress.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

