TERRA expression is regulated by the telomere-binding proteins POT-1 and POT-2 in Caenorhabditis elegans
- PMID: 37713629
- PMCID: PMC10602879
- DOI: 10.1093/nar/gkad742
TERRA expression is regulated by the telomere-binding proteins POT-1 and POT-2 in Caenorhabditis elegans
Abstract
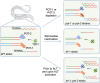
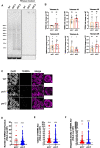
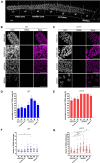
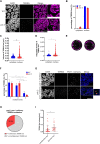
Several aspects of telomere biology are regulated by the telomeric repeat-containing RNA TERRA. While TERRA expression is conserved through evolution, species-specific mechanisms regulate its biogenesis and function. Here we report on the expression of TERRA in Caenorhabditis elegans. We show that C. elegans TERRA is regulated by the telomere-binding proteins POT-1 and POT-2 which repress TERRA in a telomere-specific manner. C. elegans TERRA transcripts are heterogeneous in length and form discrete nuclear foci, as detected by RNA FISH, in both postmitotic and germline cells; a fraction of TERRA foci localizes to telomeres. Interestingly, in germ cells, TERRA is expressed in all stages of meiotic prophase I, and it increases during pachytene, a stage in meiosis when homologous recombination is ongoing. We used the MS2-GFP system to study the spatiotemporal dynamics of single-telomere TERRA molecules. Single particle tracking revealed different types of motilities, suggesting complex dynamics of TERRA transcripts. Finally, we unveiled distinctive features of C. elegans TERRA, which is regulated by telomere shortening in a telomere-specific manner, and it is upregulated in the telomerase-deficient trt-1; pot-2 double mutant prior to activation of the alternative lengthening mechanism ALT. Interestingly, in these worms TERRA displays distinct dynamics with a higher fraction of fast-moving particles.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

