Allele-specific expression reveals genetic drivers of tissue regeneration in mice
- PMID: 37714154
- PMCID: PMC10592051
- DOI: 10.1016/j.stem.2023.08.010
Allele-specific expression reveals genetic drivers of tissue regeneration in mice
Abstract
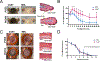
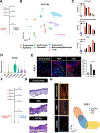
In adult mammals, skin wounds typically heal by scarring rather than through regeneration. In contrast, "super-healer" Murphy Roths Large (MRL) mice have the unusual ability to regenerate ear punch wounds; however, the molecular basis for this regeneration remains elusive. Here, in hybrid crosses between MRL and non-regenerating mice, we used allele-specific gene expression to identify cis-regulatory variation associated with ear regeneration. Analyzing three major cell populations (immune, fibroblast, and endothelial), we found that genes with cis-regulatory differences specifically in fibroblasts were associated with wound-healing pathways and also co-localized with quantitative trait loci for ear wound-healing. Ectopic treatment with one of these proteins, complement factor H (CFH), accelerated wound repair and induced regeneration in typically fibrotic wounds. Through single-cell RNA sequencing (RNA-seq), we observed that CFH treatment dramatically reduced immune cell recruitment to wounds, suggesting a potential mechanism for CFH's effect. Overall, our results provide insights into the molecular drivers of regeneration with potential clinical implications.
Keywords: fibroblasts; fibrosis; gene expression analysis; genetics; genomics; regeneration; wound healing.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

