Ustilaginoidea virens-secreted effector Uv1809 suppresses rice immunity by enhancing OsSRT2-mediated histone deacetylation
- PMID: 37715970
- PMCID: PMC10754013
- DOI: 10.1111/pbi.14174
Ustilaginoidea virens-secreted effector Uv1809 suppresses rice immunity by enhancing OsSRT2-mediated histone deacetylation
Abstract
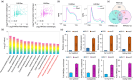
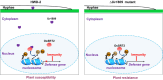
Rice false smut caused by Ustilaginoidea virens is a devastating rice (Oryza sativa) disease worldwide. However, the molecular mechanisms underlying U. virens-rice interactions are largely unknown. In this study, we identified a secreted protein, Uv1809, as a key virulence factor. Heterologous expression of Uv1809 in rice enhanced susceptibility to rice false smut and bacterial blight. Host-induced gene silencing of Uv1809 in rice enhanced resistance to U. virens, suggesting that Uv1809 inhibits rice immunity and promotes infection by U. virens. Uv1809 suppresses rice immunity by targeting and enhancing rice histone deacetylase OsSRT2-mediated histone deacetylation, thereby reducing H4K5ac and H4K8ac levels and interfering with the transcriptional activation of defence genes. CRISPR-Cas9 edited ossrt2 mutants showed no adverse effects in terms of growth and yield but displayed broad-spectrum resistance to rice pathogens, revealing a potentially valuable genetic resource for breeding disease resistance. Our study provides insight into defence mechanisms against plant pathogens that inactivate plant immunity at the epigenetic level.
Keywords: OsSRT2; Uv1809; histone acetylation; immunity; rice false smut.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
A secreted fungal effector suppresses rice immunity through host histone hypoacetylation.New Phytol. 2022 Sep;235(5):1977-1994. doi: 10.1111/nph.18265. Epub 2022 Jun 8. New Phytol. 2022. PMID: 35592995
-
The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region.Mol Plant Pathol. 2020 Apr;21(4):445-459. doi: 10.1111/mpp.12894. Epub 2020 Feb 22. Mol Plant Pathol. 2020. PMID: 32087618 Free PMC article.
-
Ustilaginoidea virens Nuclear Effector SCRE4 Suppresses Rice Immunity via Inhibiting Expression of a Positive Immune Regulator OsARF17.Int J Mol Sci. 2022 Sep 10;23(18):10527. doi: 10.3390/ijms231810527. Int J Mol Sci. 2022. PMID: 36142440 Free PMC article.
-
Ustilaginoidea virens, an emerging pathogen of rice: the dynamic interplay between the pathogen virulence strategies and host defense.Planta. 2024 Sep 11;260(4):92. doi: 10.1007/s00425-024-04523-x. Planta. 2024. PMID: 39261328 Review.
-
Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease.Mol Plant Pathol. 2016 Dec;17(9):1321-1330. doi: 10.1111/mpp.12362. Epub 2016 Apr 13. Mol Plant Pathol. 2016. PMID: 26720072 Free PMC article. Review.
Cited by
-
PtrA, Piz-t, and a novel minor-effect QTL (qBR12_3.3-4.4) collectively contribute to the durable blast-resistance of rice cultivar Tainung 84.Bot Stud. 2024 Dec 18;65(1):37. doi: 10.1186/s40529-024-00444-w. Bot Stud. 2024. PMID: 39692953 Free PMC article.
-
Selecting ChIP-seq normalization methods from the perspective of their technical conditions.Brief Bioinform. 2025 Jul 2;26(4):bbaf431. doi: 10.1093/bib/bbaf431. Brief Bioinform. 2025. PMID: 40838784 Free PMC article.
-
Advances in understanding the roles of plant HAT and HDAC in non-histone protein acetylation and deacetylation.Planta. 2024 Sep 12;260(4):93. doi: 10.1007/s00425-024-04518-8. Planta. 2024. PMID: 39264431 Review.
-
Natural variation in the pattern-triggered immunity response in plants: Investigations, implications and applications.Mol Plant Pathol. 2024 Mar;25(3):e13445. doi: 10.1111/mpp.13445. Mol Plant Pathol. 2024. PMID: 38528659 Free PMC article. Review.
-
Transcriptomic Analysis of the CNL Gene Family in the Resistant Rice Cultivar IR28 in Response to Ustilaginoidea virens Infection.Int J Mol Sci. 2024 Oct 3;25(19):10655. doi: 10.3390/ijms251910655. Int J Mol Sci. 2024. PMID: 39408984 Free PMC article.
References
-
- Azevedo, C. , Sadanandom, A. , Kitagawa, K. , Freialdenhoven, A. , Shirasu, K. and Schulze‐Lefert, P. (2002) The RAR1 interactor SGT1, an essential component of R gene‐triggered disease resistance. Science, 295, 2073–2076. - PubMed
-
- Berger, S.L. (2007) The complex language of chromatin regulation during transcription. Nature, 447, 407–412. - PubMed
-
- Chen, X. , Ding, A.B. and Zhong, X. (2020a) Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 63, 206–216. - PubMed
-
- Chen, X.Y. , Tang, J.T. , Pei, Z.X. , Liu, H. , Huang, J.B. , Luo, C.X. , Hsiang, T. et al. (2020b) The “pears and lemons” protein UvPal1 regulates development and virulence of Ustilaginoidea virens . Environ. Microbiol. 22, 5414–5432. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

