Multi-omic analysis reveals metabolic pathways that characterize right-sided colon cancer liver metastasis
- PMID: 37716465
- PMCID: PMC10620771
- DOI: 10.1016/j.canlet.2023.216384
Multi-omic analysis reveals metabolic pathways that characterize right-sided colon cancer liver metastasis
Abstract
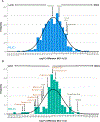
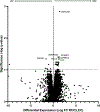
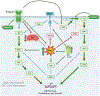
There are well demonstrated differences in tumor cell metabolism between right sided (RCC) and left sided (LCC) colon cancer, which could underlie the robust differences observed in their clinical behavior, particularly in metastatic disease. As such, we utilized liquid chromatography-mass spectrometry to perform an untargeted metabolomics analysis comparing frozen liver metastasis (LM) biobank samples derived from patients with RCC (N = 32) and LCC (N = 58) to further elucidate the unique biology of each. We also performed an untargeted RNA-seq and subsequent network analysis on samples derived from an overlapping subset of patients (RCC: N = 10; LCC: N = 18). Our biobank redemonstrates the inferior survival of patients with RCC-derived LM (P = 0.04), a well-established finding. Our metabolomic results demonstrate increased reactive oxygen species associated metabolites and bile acids in RCC. Conversely, carnitines, indicators of fatty acid oxidation, are relatively increased in LCC. The transcriptomic analysis implicates increased MEK-ERK, PI3K-AKT and Transcription Growth Factor Beta signaling in RCC LM. Our multi-omic analysis reveals several key differences in cellular physiology which taken together may be relevant to clinical differences in tumor behavior between RCC and LCC liver metastasis.
Keywords: Bile acids; EGFR; Laterality; Metabolomics; ROS; TGF-β.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

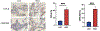
References
-
- Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, Passalacqua R, Sgroi G, Barni S, Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis, JAMA Oncol, 3 (2017) 211–219. - PubMed
-
- Yahagi M, Okabayashi K, Hasegawa H, Tsuruta M, Kitagawa Y, The Worse Prognosis of Right-Sided Compared with Left-Sided Colon Cancers: a Systematic Review and Meta-analysis, J Gastrointest Surg, 20 (2016) 648–655. - PubMed
-
- Wang ZX, Wu HX, He MM, Wang YN, Luo HY, Ding PR, Xie D, Chen G, Li YH, Wang F, Xu RH, Chemotherapy With or Without Anti-EGFR Agents in Left- and Right-Sided Metastatic Colorectal Cancer: An Updated Meta-Analysis, J Natl Compr Canc Netw, 17 (2019) 805–811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

