A novel NET-related gene signature for predicting DLBCL prognosis
- PMID: 37716978
- PMCID: PMC10504796
- DOI: 10.1186/s12967-023-04494-9
A novel NET-related gene signature for predicting DLBCL prognosis
Abstract
Background: Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignancy. Neutrophil extracellular traps (NETs) are pathogen-trapping structures in the tumor microenvironment that affect DLBCL progression. However, the predictive function of NET-related genes (NRGs) in DLBCL has received little attention. This study aimed to investigate the interaction between NRGs and the prognosis of DLBCL as well as their possible association with the immunological microenvironment.
Methods: The gene expression and clinical data of patients with DLBCL were downloaded from the Gene Expression Omnibus database. We identified 148 NRGs through the manual collection of literature. GSE10846 (n = 400, GPL570) was used as the training dataset and divided into training and testing sets in a 7:3 ratio. Univariate Cox regression analysis was used to identify overall survival (OS)-related NETs, and the least absolute shrinkage and selection operator was used to evaluate the predictive efficacy of the NRGs. Kaplan-Meier plots were used to visualize survival functions. Receiver operating characteristic (ROC) curves were used to assess the prognostic predictive ability of NRG-based features. A nomogram containing the clinical information and prognostic scores of the patients was constructed using multivariate logistic regression and Cox proportional risk regression models.
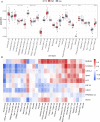
Results: We identified 36 NRGs that significantly affected patient overall survival (OS). Eight NRGs (PARVB, LYZ, PPARGC1A, HIF1A, SPP1, CDH1, S100A9, and CXCL2) were found to have excellent predictive potential for patient survival. For the 1-, 3-, and 5-year survival rates, the obtained areas under the receiver operating characteristic curve values were 0.8, 0.82, and 0.79, respectively. In the training set, patients in the high NRG risk group presented a poorer prognosis (p < 0.0001), which was validated using two external datasets (GSE11318 and GSE34171). The calibration curves of the nomogram showed that it had excellent predictive ability. Moreover, in vitro quantitative real-time PCR (qPCR) results showed that the mRNA expression levels of CXCL2, LYZ, and PARVB were significantly higher in the DLBCL group.
Conclusions: We developed a genetic risk model based on NRGs to predict the prognosis of patients with DLBCL, which may assist in the selection of treatment drugs for these patients.
Keywords: Diffuse large B-cell lymphoma; Neutrophil extracellular traps; Prognostic biomarker; Prognostic model; Tumor microenvironment.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing financial interests exist.
Figures








References
-
- International Non-Hodgkin's Lymphoma Prognostic Factors Project A predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med. 1993;329(14):987–994. - PubMed
-
- Sehn LH, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109(5):1857–1861. - PubMed
-
- Olszewski AJ, Winer ES, Castillo JJ. Validation of clinical prognostic indices for diffuse large B-cell lymphoma in the National Cancer Data Base. Cancer Causes Control. 2015;26(8):1163–1172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

