CRNDE mediated hnRNPA2B1 stability facilitates nuclear export and translation of KRAS in colorectal cancer
- PMID: 37716979
- PMCID: PMC10505224
- DOI: 10.1038/s41419-023-06137-9
CRNDE mediated hnRNPA2B1 stability facilitates nuclear export and translation of KRAS in colorectal cancer
Abstract
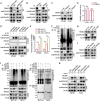
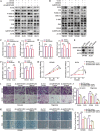
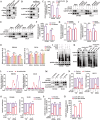
Development of colorectal cancer (CRC) involves activation of Kirsten rat sarcoma viral oncogene homolog (KRAS) signaling. However, the post-transcriptional regulation of KRAS has yet to be fully characterized. Here, we found that the colorectal neoplasia differentially expressed (CRNDE)/heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) axis was notably elevated in CRC and was strongly associated with poor prognosis of patients, while also significantly promoting CRC cell proliferation and metastasis both in vitro and in vivo. Furthermore, CRNDE maintained the stability of hnRNPA2B1 protein by inhibiting E3 ubiquitin ligase TRIM21 mediated K63 ubiquitination-dependent protein degradation. CRNDE/hnRNPA2B1 axis facilitated the nuclear export and translation of KRAS mRNA, which specifically activated the MAPK signaling pathway, eventually accelerating the malignant progression of CRC. Our findings provided insight into the regulatory network for stable hnRNPA2B1 protein expression, and the molecular mechanisms by which the CRNDE/hnRNPA2B1 axis mediated KRAS nucleocytoplasmic transport and translation, deeply underscoring the bright future of hnRNPA2B1 as a promising biomarker and therapeutic target for CRC. By hindering hnRNPA2B1 from binding to the E3 ubiquitin ligase TRIM21, whose mediated ubiquitin-dependent degradation was thereby inhibited, CRNDE protected the stability of hnRNPA2B1's high protein expression in CRC. Supported by the high level of the oncogenic molecule CRNDE, hnRNPA2B1 bound to KRAS mRNA and promoted KRAS mRNA nucleus export to enter the ribosomal translation program, subsequently activating the MAPK signaling pathway and ultimately accelerating the malignant progression of CRC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways.Cell Death Dis. 2017 Jun 8;8(6):e2862. doi: 10.1038/cddis.2017.258. Cell Death Dis. 2017. PMID: 28594403 Free PMC article.
-
Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells.Gastroenterology. 2014 Oct;147(4):882-892.e8. doi: 10.1053/j.gastro.2014.06.041. Epub 2014 Jul 3. Gastroenterology. 2014. PMID: 24998203
-
MicroRNA‑337 inhibits colorectal cancer progression by directly targeting KRAS and suppressing the AKT and ERK pathways.Oncol Rep. 2017 Nov;38(5):3187-3196. doi: 10.3892/or.2017.5997. Epub 2017 Sep 25. Oncol Rep. 2017. Retraction in: Oncol Rep. 2022 Oct;48(4):179. doi: 10.3892/or.2022.8394. PMID: 29048669 Retracted.
-
Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer.Mol Cancer. 2021 Nov 6;20(1):143. doi: 10.1186/s12943-021-01441-4. Mol Cancer. 2021. PMID: 34742312 Free PMC article. Review.
-
The current understanding on the impact of KRAS on colorectal cancer.Biomed Pharmacother. 2021 Aug;140:111717. doi: 10.1016/j.biopha.2021.111717. Epub 2021 May 24. Biomed Pharmacother. 2021. PMID: 34044280 Review.
Cited by
-
HNRNPA2B1: a novel target in pulmonary arterial hypertension.Front Cardiovasc Med. 2025 Jul 9;12:1497938. doi: 10.3389/fcvm.2025.1497938. eCollection 2025. Front Cardiovasc Med. 2025. PMID: 40703632 Free PMC article. Review.
-
Prolyl 4-hydroxylase subunit alpha-2 acts as a TRIM21 ubiquitination substrate to promote papillary thyroid cancer progression via the glycolytic pathway.Cell Death Dis. 2025 May 17;16(1):395. doi: 10.1038/s41419-025-07702-0. Cell Death Dis. 2025. PMID: 40379610 Free PMC article.
-
EGFR-induced lncRNA TRIDENT promotes drug resistance in non-small cell lung cancer via phospho-TRIM28-mediated DNA damage repair.Proc Natl Acad Sci U S A. 2025 Mar 11;122(10):e2415389122. doi: 10.1073/pnas.2415389122. Epub 2025 Mar 3. Proc Natl Acad Sci U S A. 2025. PMID: 40030013 Free PMC article.
-
hnRNPA2B1 drives colorectal cancer progression via the circCDYL/EIF4A3/PHF8 axis.Kaohsiung J Med Sci. 2025 Mar;41(3):e12943. doi: 10.1002/kjm2.12943. Epub 2025 Jan 15. Kaohsiung J Med Sci. 2025. PMID: 39810713 Free PMC article.
-
Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC-A New Opportunity to Predict Therapy Response.Cancers (Basel). 2023 Nov 28;15(23):5623. doi: 10.3390/cancers15235623. Cancers (Basel). 2023. PMID: 38067326 Free PMC article.
References
-
- Messersmith WA. NCCN Guidelines Updates: Management of Metastatic Colorectal Cancer. J Natl Compr Canc Netw. 2019;17:599–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

