Ustilago maydis PR-1-like protein has evolved two distinct domains for dual virulence activities
- PMID: 37716995
- PMCID: PMC10505147
- DOI: 10.1038/s41467-023-41459-4
Ustilago maydis PR-1-like protein has evolved two distinct domains for dual virulence activities
Abstract
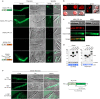
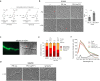
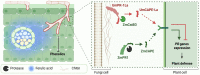
The diversification of effector function, driven by a co-evolutionary arms race, enables pathogens to establish compatible interactions with hosts. Structurally conserved plant pathogenesis-related PR-1 and PR-1-like (PR-1L) proteins are involved in plant defense and fungal virulence, respectively. It is unclear how fungal PR-1L counters plant defense. Here, we show that Ustilago maydis UmPR-1La and yeast ScPRY1, with conserved phenolic resistance functions, are Ser/Thr-rich region mediated cell-surface localization proteins. However, UmPR-1La has gained specialized activity in sensing phenolics and eliciting hyphal-like formation to guide fungal growth in plants. Additionally, U. maydis hijacks maize cathepsin B-like 3 (CatB3) to release functional CAPE-like peptides by cleaving UmPR-1La's conserved CNYD motif, subverting plant CAPE-primed immunity and promoting fungal virulence. Surprisingly, CatB3 avoids cleavage of plant PR-1s, despite the presence of the same conserved CNYD motif. Our work highlights that UmPR-1La has acquired additional dual roles to suppress plant defense and sustain the infection process of fungal pathogens.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The functionally conserved effector Sta1 is a fungal cell wall protein required for virulence in Ustilago maydis.New Phytol. 2020 Jul;227(1):185-199. doi: 10.1111/nph.16508. Epub 2020 Mar 25. New Phytol. 2020. PMID: 32112567
-
Cross-species analysis between the maize smut fungi Ustilago maydis and Sporisorium reilianum highlights the role of transcriptional change of effector orthologs for virulence and disease.New Phytol. 2021 Oct;232(2):719-733. doi: 10.1111/nph.17625. Epub 2021 Aug 6. New Phytol. 2021. PMID: 34270791
-
Dual function of a secreted fungalysin metalloprotease in Ustilago maydis.New Phytol. 2018 Oct;220(1):249-261. doi: 10.1111/nph.15265. Epub 2018 Jun 19. New Phytol. 2018. PMID: 29916208
-
Ustilago maydis effectors and their impact on virulence.Nat Rev Microbiol. 2017 Jul;15(7):409-421. doi: 10.1038/nrmicro.2017.33. Epub 2017 May 8. Nat Rev Microbiol. 2017. PMID: 28479603 Review.
-
Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis--similar inputs, different outputs.Curr Opin Microbiol. 2001 Apr;4(2):214-21. doi: 10.1016/s1369-5274(00)00191-0. Curr Opin Microbiol. 2001. PMID: 11282479 Review.
Cited by
-
How plants manage pathogen infection.EMBO Rep. 2024 Jan;25(1):31-44. doi: 10.1038/s44319-023-00023-3. Epub 2023 Dec 19. EMBO Rep. 2024. PMID: 38177909 Free PMC article. Review.
-
Foliar Pine Pathogens From Different Kingdoms Share Defence-Eliciting Effector Proteins.Mol Plant Pathol. 2025 Mar;26(3):e70065. doi: 10.1111/mpp.70065. Mol Plant Pathol. 2025. PMID: 40025648 Free PMC article.
-
Plant Defense Proteins: Recent Discoveries and Applications.Plants (Basel). 2025 Jul 6;14(13):2069. doi: 10.3390/plants14132069. Plants (Basel). 2025. PMID: 40648078 Free PMC article. Review.
-
In silico characterization, physiochemical analysis, and antifungal evaluation of the Zea mays PR-1 protein.Sci Rep. 2025 Aug 27;15(1):31523. doi: 10.1038/s41598-025-16772-1. Sci Rep. 2025. PMID: 40866506 Free PMC article.
-
Identification of CAP genes in finger lime (Citrus australasica) and their role in plant responses to abiotic and biotic stress.Sci Rep. 2024 Nov 28;14(1):29557. doi: 10.1038/s41598-024-80868-3. Sci Rep. 2024. PMID: 39632943 Free PMC article.
References
-
- Gibbs GM, Roelants K, O’Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008;29:865–897. - PubMed
-
- Schneiter R, Di Pietro A. The CAP protein superfamily: function in sterol export and fungal virulence. Biomol. Concepts. 2013;4:519–525. - PubMed
-
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. - PubMed
-
- Breen S, Williams SJ, Outram M, Kobe B, Solomon PS. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017;22:871–879. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

