Structure adaptation in Omicron SARS-CoV-2/hACE2: Biophysical origins of evolutionary driving forces
- PMID: 37717145
- PMCID: PMC10624932
- DOI: 10.1016/j.bpj.2023.09.003
Structure adaptation in Omicron SARS-CoV-2/hACE2: Biophysical origins of evolutionary driving forces
Abstract
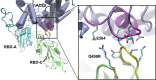
Since its emergence, the COVID-19 threat has been sustained by a series of transmission waves initiated by new variants of the SARS-CoV-2 virus. Some of these arise with higher transmissivity and/or increased disease severity. Here, we use molecular dynamics simulations to examine the modulation of the fundamental interactions between the receptor binding domain (RBD) of the spike glycoprotein and the host cell receptor (human angiotensin-converting enzyme 2 [hACE2]) arising from Omicron variant mutations (BA.1 and BA.2) relative to the original wild-type strain. Our key findings are that glycans play a vital role at the RBD···hACE2 interface for the Omicrons, and the interplay between glycans and sequence mutations leads to enhanced binding. We find significant structural differences in the complexes, which overall bring the spike protein and its receptor into closer proximity. These are consistent with and attributed to the higher positive charge on the RBD conferred by BA.1 and BA.2 mutations relative to the wild-type. However, further differences between subvariants BA.1 and BA.2 (which have equivalent RBD charges) are also evident: mutations reduce interdomain interactions between the up chain and its clockwise neighbor chain in particular for the latter, resulting in enhanced flexibility for BA.2. Consequently, we see occurrence of additional close contacts in one replica of BA.2, which include binding to hACE2 by a second RBD in addition to the up chain. Although this motif is not seen in BA.1, we find that the Omicrons can directly/indirectly bind a down-RBD to hACE2 through glycans: the role of the glycan on N90 of hACE2 switches from inhibiting to facilitating the binding to Omicron spike protein via glycan-protein lateral interactions. These structural and electrostatic differences offer further insight into the mechanisms by which viral mutations modulate host cell binding and provide a biophysical basis for evolutionary driving forces.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






Similar articles
-
Improved Binding Affinity of Omicron's Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence.Int J Mol Sci. 2022 Mar 21;23(6):3409. doi: 10.3390/ijms23063409. Int J Mol Sci. 2022. PMID: 35328828 Free PMC article.
-
Investigating the role of glycans in Omicron sub-lineages XBB.1.5 and XBB.1.16 binding to host receptor using molecular dynamics and binding free energy calculations.J Comput Aided Mol Des. 2023 Nov;37(11):551-563. doi: 10.1007/s10822-023-00526-0. Epub 2023 Aug 5. J Comput Aided Mol Des. 2023. PMID: 37542610
-
Structural and Energetic Insights into SARS-CoV-2 Evolution: Analysis of hACE2-RBD Binding in Wild-Type, Delta, and Omicron Subvariants.Int J Mol Sci. 2025 Apr 17;26(8):3776. doi: 10.3390/ijms26083776. Int J Mol Sci. 2025. PMID: 40332432 Free PMC article.
-
Surface charge changes in spike RBD mutations of SARS-CoV-2 and its variant strains alter the virus evasiveness via HSPGs: A review and mechanistic hypothesis.Front Public Health. 2022 Aug 24;10:952916. doi: 10.3389/fpubh.2022.952916. eCollection 2022. Front Public Health. 2022. PMID: 36091499 Free PMC article. Review.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
Cited by
-
Balancing stability and function: impact of the surface charge of SARS-CoV-2 Omicron spike protein.Npj Viruses. 2025 Apr 1;3(1):23. doi: 10.1038/s44298-025-00104-1. Npj Viruses. 2025. PMID: 40295844 Free PMC article. Review.
-
Viral entry mechanisms: the role of molecular simulation in unlocking a key step in viral infections.FEBS Open Bio. 2025 Feb;15(2):269-284. doi: 10.1002/2211-5463.13908. Epub 2024 Oct 14. FEBS Open Bio. 2025. PMID: 39402013 Free PMC article. Review.
-
Managing and treating COVID-19 in patients with hematological malignancies: a narrative review and expert insights.Clin Exp Med. 2024 Jun 4;24(1):119. doi: 10.1007/s10238-024-01381-5. Clin Exp Med. 2024. PMID: 38833206 Free PMC article. Review.
References
-
- Planas D., Veyer D., et al. Schwartz O. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature. 2021;596:276–280. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

