Comparative expression analysis of sucrose phosphate synthase gene family in a low and high sucrose Pakistani sugarcane cultivars
- PMID: 37719124
- PMCID: PMC10503496
- DOI: 10.7717/peerj.15832
Comparative expression analysis of sucrose phosphate synthase gene family in a low and high sucrose Pakistani sugarcane cultivars
Abstract
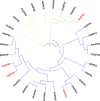
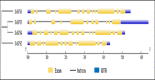
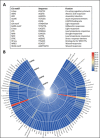
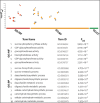
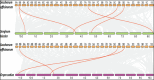
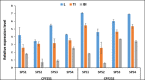
Sugarcane is the world's largest cultivated crop by biomass and is the main source of sugar and biofuel. Sucrose phosphate synthase (SPS) enzymes are directly involved in the synthesis of sucrose. Here, we analyzed and compared one of the important gene families involved in sucrose metabolism in a high and low sucrose sugarcane cultivar. A comprehensive in silico analysis of the SoSPS family displayed their phylogenetic relationship, gene and protein structure, miRNA targets, protein interaction network (PPI), gene ontology and collinearity. This was followed by a spatial expression analysis in two different sugarcane varieties. The phylogenetic reconstruction distributed AtSPS, ZmSPS, OsSPS, SoSPS and SbSPS into three main groups (A, B, C). The regulatory region of SoSPS genes carries ABRE, ARE, G-box, and MYC as the most dominant cis-regulatory elements. The PPI analysis predicted a total of 14 unique proteins interacting with SPS. The predominant expression of SPS in chloroplast clearly indicates that they are the most active in the organelle which is the hub of photosynthesis. Similarly, gene ontology attributed SPS to sucrose phosphate synthase and glucosyl transferase molecular functions, as well as sucrose biosynthetic and disaccharide biological processes. Overall, the expression of SPS in CPF252 (high sucrose variety) was higher in leaf and culm as compared to that of CPF 251 (low sucrose variety). In brief, this study adds to the present literature about sugarcane, sucrose metabolism and role of SPS in sucrose metabolism thereby opening up further avenues of research in crop improvement.
Keywords: Amino acid; Arabidopsis; Gene; High level; Physiochemical; Protein; Sucrose; Sucrose phosphate synthase enzymes; Sugar cane; synthesis.
©2023 Niazi et al.
Conflict of interest statement
Jiban Shrestha is an Academic Editor for PeerJ.
Figures



Similar articles
-
Comparative analysis of sucrose phosphate synthase (SPS) gene family between Saccharum officinarum and Saccharum spontaneum.BMC Plant Biol. 2020 Sep 14;20(1):422. doi: 10.1186/s12870-020-02599-7. BMC Plant Biol. 2020. PMID: 32928111 Free PMC article.
-
Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars.Plant Biol (Stuttg). 2011 Mar;13(2):325-32. doi: 10.1111/j.1438-8677.2010.00379.x. Epub 2010 Aug 26. Plant Biol (Stuttg). 2011. PMID: 21309979
-
Sucrose-phosphate phosphatase from sugarcane reveals an ancestral tandem duplication.BMC Plant Biol. 2021 Jan 7;21(1):23. doi: 10.1186/s12870-020-02795-5. BMC Plant Biol. 2021. PMID: 33413115 Free PMC article.
-
Sucrose Phosphate Synthase Genes in Plants: Its Role and Practice for Crop Improvement.J Agric Food Chem. 2024 Aug 21;72(33):18335-18346. doi: 10.1021/acs.jafc.4c05068. Epub 2024 Aug 12. J Agric Food Chem. 2024. PMID: 39134474 Review.
-
Identification of UDP-glucose binding site in glycosyltransferase domain of sucrose phosphate synthase from sugarcane (Saccharum officinarum) by structure-based site-directed mutagenesis.Biophys Rev. 2018 Apr;10(2):293-298. doi: 10.1007/s12551-017-0360-9. Epub 2017 Dec 8. Biophys Rev. 2018. PMID: 29222806 Free PMC article. Review.
Cited by
-
Proteomic Analysis of Plants with Binding Immunoglobulin Protein Overexpression Reveals Mechanisms Related to Defense Against Moniliophthora perniciosa.Plants (Basel). 2025 Feb 7;14(4):503. doi: 10.3390/plants14040503. Plants (Basel). 2025. PMID: 40006761 Free PMC article.
References
-
- Aguilar-Rivera N, Rodríguez LDA, Enríquez RV, Castillo MA, Herrera SA. The Mexican sugarcane industry: overview, constraints, current status and long-term trends. Sugar Tech. 2012;14:207–222. doi: 10.1007/s12355-012-0151-3. - DOI
-
- Almadanim MC, Alexandre BM, Rosa MTG, Sapeta H, Leitão AE, Ramalho JC, Lam TT, Negrão S, Abreu IA, Oliveira MM. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant, Cell & Environment. 2017;40:1197–1213. doi: 10.1111/pce.12916. - DOI - PubMed
-
- Batta SK, Singh R. Sucrose metabolism in sugar cane grown under varying climatic conditions: synthesis and storage of sucrose in relation to the activities of sucrose synthase, sucrose-phosphate synthase and invertase. Phytochemistry. 1986;25:2431–2437. doi: 10.1016/S0031-9422(00)84484-2. - DOI
-
- Bhat KV, Mondal TK, Gaikwad AB, Kole PR, Chandel G, Mohapatra T. Genome-wide identification of drought-responsive miRNAs in grass pea (Lathyrus sativus L) Plant Gene. 2020;21:100210. doi: 10.1016/j.plgene.2019.100210. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

