Identification of novel candidate loci and genes for seed vigor-related traits in upland cotton (Gossypium hirsutum L.) via GWAS
- PMID: 37719213
- PMCID: PMC10503134
- DOI: 10.3389/fpls.2023.1254365
Identification of novel candidate loci and genes for seed vigor-related traits in upland cotton (Gossypium hirsutum L.) via GWAS
Abstract
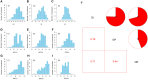
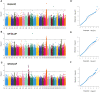
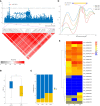
Seed vigor (SV) is a crucial trait determining the quality of crop seeds. Currently, over 80% of China's cotton-planting area is in Xinjiang Province, where a fully mechanized planting model is adopted, accounting for more than 90% of the total fiber production. Therefore, identifying SV-related loci and genes is crucial for improving cotton yield in Xinjiang. In this study, three seed vigor-related traits, including germination potential, germination rate, and germination index, were investigated across three environments in a panel of 355 diverse accessions based on 2,261,854 high-quality single-nucleotide polymorphisms (SNPs). A total of 26 significant SNPs were detected and divided into six quantitative trait locus regions, including 121 predicted candidate genes. By combining gene expression, gene annotation, and haplotype analysis, two novel candidate genes (Ghir_A09G002730 and Ghir_D03G009280) within qGR-A09-1 and qGI/GP/GR-D03-3 were associated with vigor-related traits, and Ghir_A09G002730 was found to be involved in artificial selection during cotton breeding by population genetic analysis. Thus, understanding the genetic mechanisms underlying seed vigor-related traits in cotton could help increase the efficiency of direct seeding by molecular marker-assisted selection breeding.
Keywords: GWAS; candidate genes; germination rate; seed vigor; upland cotton.
Copyright © 2023 Li, Hu, Wang, Zhao, You, Liu, Wang, Yan, Zhao, Huang, Yu and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andrews S. (2010). “FastQC: a quality control tool for high throughput sequence data,” Babraham Bioinformatics (United Kingdom: Babraham Bioinformatics, Babraham Institute, Cambridge; ).
LinkOut - more resources
Full Text Sources

