Active site determination of novel plant versatile peroxidase extracted from Citrus sinensis and bioconversion of β-naphthol
- PMID: 37719748
- PMCID: PMC10501043
- DOI: 10.1007/s13205-023-03758-x
Active site determination of novel plant versatile peroxidase extracted from Citrus sinensis and bioconversion of β-naphthol
Abstract
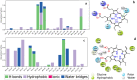
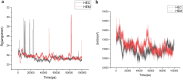
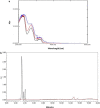
A ligninolytic peroxidase called versatile peroxidase, VP, (EC 1.11.1.16) is an iron-containing metalloenzyme. The most distinctive feature of this enzyme is its composite molecular framework, which combines lignin peroxidase's capacity to oxidize compounds with high-redox potential with manganese peroxidase's capacity to oxidize Mn2+ to Mn3+. In this study, we have extracted amino acid sequences from the Citrus sinensis source and subjected them to various computation tools to visualize the insight secondary and 3D structure, physicochemical properties, and validation of the structure which have not been studied so far to further investigate the catalytic efficiency and effectiveness of VP. The binding energies of HEME and HEME C (HEC) ligands with produced PDB (6rqf.1. A) have been also assessed, analyzed, and confirmed utilizing AutoDock. Binding energies were calculated using the AutoDock and validated by MD simulation using SCHRODINGER DESMOND. Most stable confirmation was achieved through a protein-ligand interaction study. Bio-technological use of VP in the biotransformation of β-naphthol has also been studied. The findings in the current study will have a substantial impact on proteomics, biochemistry, biotechnology, and possible uses of versatile peroxidase in the bio-remediation of different toxic organic compounds.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-023-03758-x.
Keywords: 3D structure; In silico; Metalloenzyme; Physicochemical; Protein–protein interaction; Versatile peroxidase.
© King Abdulaziz City for Science and Technology 2023. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that there is no conflict of interest.
Figures



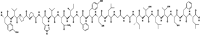



References
-
- Bathula R, Lanka G, Muddagoni N, et al. Identification of potential Aurora kinase-C protein inhibitors: an amalgamation of energy minimization, virtual screening, prime MMGBSA and AutoDock. J Biomol Struct Dyn. 2019;2:2. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

