Development and validation of a deep learning-based automatic segmentation model for assessing intracranial volume: comparison with NeuroQuant, FreeSurfer, and SynthSeg
- PMID: 37719763
- PMCID: PMC10503131
- DOI: 10.3389/fneur.2023.1221892
Development and validation of a deep learning-based automatic segmentation model for assessing intracranial volume: comparison with NeuroQuant, FreeSurfer, and SynthSeg
Erratum in
-
Corrigendum: Development and validation of a deep learning-based automatic segmentation model for assessing intracranial volume: comparison with NeuroQuant, FreeSurfer, and SynthSeg.Front Neurol. 2023 Nov 29;14:1334962. doi: 10.3389/fneur.2023.1334962. eCollection 2023. Front Neurol. 2023. PMID: 38093753 Free PMC article.
Abstract
Background and purpose: To develop and validate a deep learning-based automatic segmentation model for assessing intracranial volume (ICV) and to compare the accuracy determined by NeuroQuant (NQ), FreeSurfer (FS), and SynthSeg.
Materials and methods: This retrospective study included 60 subjects [30 Alzheimer's disease (AD), 21 mild cognitive impairment (MCI), 9 cognitively normal (CN)] from a single tertiary hospital for the training and validation group (50:10). The test group included 40 subjects (20 AD, 10 MCI, 10 CN) from the ADNI dataset. We propose a robust ICV segmentation model based on the foundational 2D UNet architecture trained with four types of input images (both single and multimodality using scaled or unscaled T1-weighted and T2-FLAIR MR images). To compare with our model, NQ, FS, and SynthSeg were also utilized in the test group. We evaluated the model performance by measuring the Dice similarity coefficient (DSC) and average volume difference.
Results: The single-modality model trained with scaled T1-weighted images showed excellent performance with a DSC of 0.989 ± 0.002 and an average volume difference of 0.46% ± 0.38%. Our multimodality model trained with both unscaled T1-weighted and T2-FLAIR images showed similar performance with a DSC of 0.988 ± 0.002 and an average volume difference of 0.47% ± 0.35%. The overall average volume difference with our model showed relatively higher accuracy than NQ (2.15% ± 1.72%), FS (3.69% ± 2.93%), and SynthSeg (1.88% ± 1.18%). Furthermore, our model outperformed the three others in each subgroup of patients with AD, MCI, and CN subjects.
Conclusion: Our deep learning-based automatic ICV segmentation model showed excellent performance for the automatic evaluation of ICV.
Keywords: artificial intelligence; brain; deep learning; intracranial volume segmentation; neurodegenerative disease.
Copyright © 2023 Suh, Jung, Suh, Kim, Oh, Heo, Shim, Lim, Lee, Kim and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

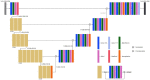

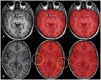
References
-
- Suh CH, Shim WH, Kim SJ, Roh JH, Lee JH, Kim MJ, et al. . Development and validation of a deep learning-based automatic brain segmentation and classification algorithm for Alzheimer disease using 3D T1-weighted volumetric images. AJNR Am J Neuroradiol. (2020) 41:2227–34. doi: 10.3174/ajnr.A6848, PMID: - DOI - PMC - PubMed
-
- Hansen TI, Brezova V, Eikenes L, Haberg A, Vangberg TR. How does the accuracy of intracranial volume measurements affect normalized brain volumes? Sample size estimates based on 966 subjects from the HUNT MRI cohort. AJNR Am J Neuroradiol. (2015) 36:1450–6. doi: 10.3174/ajnr.A4299, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

