This is a preprint.
Temporal dynamics and genomic programming of plasma cell fates
- PMID: 37720050
- PMCID: PMC10503833
- DOI: 10.21203/rs.3.rs-3296446/v1
Temporal dynamics and genomic programming of plasma cell fates
Update in
-
Temporal dynamics and genomic programming of plasma cell fates.Nat Immunol. 2024 Jun;25(6):1097-1109. doi: 10.1038/s41590-024-01831-y. Epub 2024 May 2. Nat Immunol. 2024. PMID: 38698087
Abstract
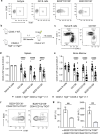
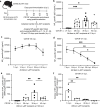
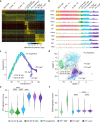
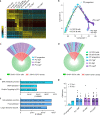
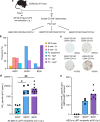
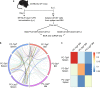
Affinity-matured plasma cells (PCs) of varying lifespans are generated through a germinal center (GC) response. The developmental dynamics and genomic programs of antigen-specific PC precursors remain to be elucidated. Using a model antigen, we demonstrate biphasic generation of PC precursors, with those generating long-lived bone marrow PCs preferentially produced in the late phase of GC response. Clonal tracing using scRNA-seq+BCR-seq in spleen and bone marrow compartments, coupled with adoptive transfer experiments, reveal a novel PC transition state that gives rise to functionally competent PC precursors. The latter undergo clonal expansion, dependent on inducible expression of TIGIT. We propose a model for the proliferation and programming of precursors of long-lived PCs, based on extended antigen encounters followed by reduced antigen availability.
Keywords: BCRseq; Plasma cell precursors; clonal tracking; durability of PCs; scRNAseq; temporal PC dynamics.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Temporal dynamics and genomic programming of plasma cell fates.Nat Immunol. 2024 Jun;25(6):1097-1109. doi: 10.1038/s41590-024-01831-y. Epub 2024 May 2. Nat Immunol. 2024. PMID: 38698087
-
Cell fate dynamics and genomic programming of plasma cell precursors.Immunol Rev. 2021 Sep;303(1):62-71. doi: 10.1111/imr.13010. Epub 2021 Jun 30. Immunol Rev. 2021. PMID: 34195999 Review.
-
Quantification of plasma cell dynamics using mathematical modelling.R Soc Open Sci. 2018 Jan 24;5(1):170759. doi: 10.1098/rsos.170759. eCollection 2018 Jan. R Soc Open Sci. 2018. PMID: 29410799 Free PMC article.
-
Short-lived and long-lived bone marrow plasma cells are derived from a novel precursor population.J Exp Med. 2002 Mar 18;195(6):737-45. doi: 10.1084/jem.20011626. J Exp Med. 2002. PMID: 11901199 Free PMC article.
-
Germinal center-dependent and -independent immune responses of tumor-infiltrating B cells in human cancers.Cell Mol Immunol. 2023 Sep;20(9):1040-1050. doi: 10.1038/s41423-023-01060-7. Epub 2023 Jul 7. Cell Mol Immunol. 2023. PMID: 37419983 Free PMC article. Review.
References
-
- Benner R., van Oudenaren A. & de Ruiter H. Antibody formation in mouse bone marrow: IX. Peripheral lymphoid organs are involved in the initiation of bone marrow antibody formation. Cellular Immunology 34, 125–137 (1977). - PubMed
-
- Smith K.G.C., Hewitson T.D., Nossal G.J.V. & Tarlinton D.M. The phenotype and fate of the antibody-forming cells of the splenic foci. European Journal of Immunology 26, 444–448 (1996). - PubMed
-
- Manz R.A., Thiel A. & Radbruch A. Lifetime of plasma cells in the bone marrow. Nature 388, 133–134 (1997). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

