Prophylactic feeding of neomycin to Holstein calves alters gut microbiota, bile acid metabolism, and expression of genes involved in immunometabolic regulation
- PMID: 37720145
- PMCID: PMC10500837
- DOI: 10.3389/fmicb.2023.1210142
Prophylactic feeding of neomycin to Holstein calves alters gut microbiota, bile acid metabolism, and expression of genes involved in immunometabolic regulation
Abstract
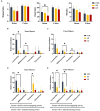
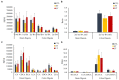
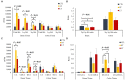
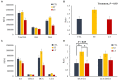
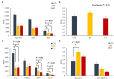
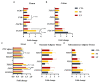
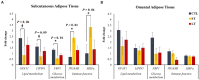
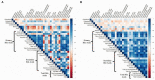
The objective of this study was to evaluate the effects of prophylactic neomycin administration on Holstein bull calves' intestinal microbiota, bile acid (BA) metabolism, and transcript abundance of genes related to BA metabolism. A total of 36 calves were blocked by body weight and assigned to either non-medicated milk replacer (CTL), or neomycin for 14 days (ST) or 28 days (LT) in their milk replacer. At the end of the study, calves were euthanized to collect tissue and digesta samples from the gastrointestinal tract, liver, and adipose tissue for analysis of intestinal microbial diversity, bile acid concentration and profile in various body tissues, and gene expression related to bile acid, lipid, carbohydrate metabolism, and inflammation. Calves that received prophylactic administration of neomycin for 28 d (LT) had reduced species richness (chao1 index), and tended to have reduced phylogenetic diversity in the ileum tissue. The relative abundance of Lactobacillus, and Bifidobacterium in ileum and colon digesta were decreased in LT compared with CTL. Concentrations of primary, secondary, and total BA were increased by ST in ileal tissue. In plasma, ST and LT treatments had lower concentrations of secondary BA. Gene expression of the BA receptor FXR was increased in ileum and liver by LT compared to CTL. The expression of FXR and TGR5 in the liver was increased in the ST group compared with CTL, and in adipose tissue, 5 genes related to triglyceride, gluconeogenesis, and immune activation were differentially expressed between CTL and ST. In conclusion, we provide evidence that prophylactic administration of neomycin leads to aberrant changes in BA concentration and profile in different compartments of the enterohepatic system through a process that possibly entails antimicrobial disruption of key bacterial groups, which persists even after cessation of neomycin administration. Additionally, we uncovered an apparent link between dysregulated BA metabolism and changes in lipid metabolism and immune activation in adipose tissue and liver.
Keywords: antibiotics; bile acids; dysbiosis; dyslipidemia; metabolism.
Copyright © 2023 Cangiano, Ipharraguerre, Guan, Buss, Amorin-Hegedus, Chirivi, Contreras and Steele.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
LinkOut - more resources
Full Text Sources

