Advances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes
- PMID: 37720320
- PMCID: PMC10502170
- DOI: 10.3389/fbioe.2023.1215466
Advances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes
Abstract
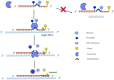
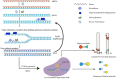
The rapid diagnosis of pathogenic infections plays a vital role in disease prevention, control, and public health safety. Recombinase-aided amplification (RAA) is an innovative isothermal nucleic acid amplification technology capable of fast DNA or RNA amplification at low temperatures. RAA offers advantages such as simplicity, speed, precision, energy efficiency, and convenient operation. This technology relies on four essential components: recombinase, single-stranded DNA-binding protein (SSB), DNA polymerase, and deoxyribonucleoside triphosphates, which collectively replace the laborious thermal cycling process of traditional polymerase chain reaction (PCR). In recent years, the CRISPR-Cas (clustered regularly interspaced short palindromic repeats-associated proteins) system, a groundbreaking genome engineering tool, has garnered widespread attention across biotechnology, agriculture, and medicine. Increasingly, researchers have integrated the recombinase polymerase amplification system (or RAA system) with CRISPR technology, enabling more convenient and intuitive determination of detection results. This integration has significantly expanded the application of RAA in pathogen detection. The step-by-step operation of these two systems has been successfully employed for molecular diagnosis of pathogenic microbes, while the single-tube one-step method holds promise for efficient pathogen detection. This paper provides a comprehensive review of RAA combined with CRISPR-Cas and its applications in pathogen detection, aiming to serve as a valuable reference for further research in related fields.
Keywords: clustered regularly interspaced short palindromic repeats associated proteins; pathogenic microbes; quick detection; recombinase-aided amplification; specific diagnosis.
Copyright © 2023 Li, Zhu, Zhang, Ren, He, Zhou, Yin, Wang, Zhong, Wang, Xiao, Zhu, Yin and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

