Detection of Frog Virus 3 by Integrating RPA-CRISPR/Cas12a-SPM with Deep Learning
- PMID: 37720737
- PMCID: PMC10500685
- DOI: 10.1021/acsomega.3c02929
Detection of Frog Virus 3 by Integrating RPA-CRISPR/Cas12a-SPM with Deep Learning
Abstract
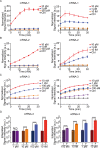
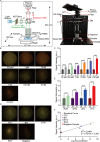
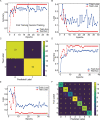
A fast, easy-to-implement, highly sensitive, and point-of-care (POC) detection system for frog virus 3 (FV3) is proposed. Combining recombinase polymerase amplification (RPA) and CRISPR/Cas12a, a limit of detection (LoD) of 100 aM (60.2 copies/μL) is achieved by optimizing RPA primers and CRISPR RNAs (crRNAs). For POC detection, smartphone microscopy is implemented, and an LoD of 10 aM is achieved in 40 min. The proposed system detects four positive animal-derived samples with a quantitation cycle (Cq) value of quantitative PCR (qPCR) in the range of 13 to 32. In addition, deep learning models are deployed for binary classification (positive or negative samples) and multiclass classification (different concentrations of FV3 and negative samples), achieving 100 and 98.75% accuracy, respectively. Without temperature regulation and expensive equipment, the proposed RPA-CRISPR/Cas12a combined with smartphone readouts and artificial-intelligence-assisted classification showcases the great potential for FV3 detection, specifically POC detection of DNA virus.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
DNA Virus Detection System Based on RPA-CRISPR/Cas12a-SPM and Deep Learning.J Vis Exp. 2024 May 10;(207). doi: 10.3791/64833. J Vis Exp. 2024. PMID: 38801262
-
One-pot platform for rapid detecting virus utilizing recombinase polymerase amplification and CRISPR/Cas12a.Appl Microbiol Biotechnol. 2022 Jun;106(12):4607-4616. doi: 10.1007/s00253-022-12015-9. Epub 2022 Jun 16. Appl Microbiol Biotechnol. 2022. PMID: 35708748 Free PMC article.
-
A rapid and high-throughput Helicobacter pylori RPA-CRISPR/Cas12a-based nucleic acid detection system.Clin Chim Acta. 2023 Feb 1;540:117201. doi: 10.1016/j.cca.2022.12.013. Epub 2022 Dec 23. Clin Chim Acta. 2023. PMID: 36572137
-
Development of a RPA-CRISPR/Cas12a based rapid visual detection assay for Porcine Parvovirus 7.Front Vet Sci. 2024 Sep 9;11:1440769. doi: 10.3389/fvets.2024.1440769. eCollection 2024. Front Vet Sci. 2024. PMID: 39315085 Free PMC article.
-
M-CDC: Magnetic pull-down-assisted colorimetric method based on the CRISPR/Cas12a system.Methods. 2022 Jul;203:259-267. doi: 10.1016/j.ymeth.2021.11.009. Epub 2021 Nov 20. Methods. 2022. PMID: 34813932 Review.
Cited by
-
Unraveling the influence of CRISPR/Cas13a reaction components on enhancing trans-cleavage activity for ultrasensitive on-chip RNA detection.Mikrochim Acta. 2024 Jul 17;191(8):466. doi: 10.1007/s00604-024-06545-4. Mikrochim Acta. 2024. PMID: 39017814
-
Mimicking an in cellulo environment for enzyme-free paper-based nucleic acid tests at the point of care.bioRxiv [Preprint]. 2024 Feb 29:2024.02.27.582375. doi: 10.1101/2024.02.27.582375. bioRxiv. 2024. Update in: ACS Sens. 2024 Oct 25;9(10):5069-5080. doi: 10.1021/acssensors.4c00539. PMID: 38464301 Free PMC article. Updated. Preprint.
-
Mimicking a Cellular Crowding Environment for Enzyme-Free Paper-Based Nucleic Acid Tests at the Point of Care.ACS Sens. 2024 Oct 25;9(10):5069-5080. doi: 10.1021/acssensors.4c00539. Epub 2024 Sep 30. ACS Sens. 2024. PMID: 39344686 Free PMC article.
References
-
- Price S. J.; Garner T. W.; Nichols R. A.; Balloux F.; Ayres C.; Mora-Cabello D. A. A.; Bosch J. Collapse of amphibian communities due to an introduced Ranavirus. Curr. Biol. 2014, 24 (21), 2586–2591. - PubMed
LinkOut - more resources
Full Text Sources

