Discovery of Novel Naphthoquinone-Chalcone Hybrids as Potent FGFR1 Tyrosine Kinase Inhibitors: Synthesis, Biological Evaluation, and Molecular Modeling
- PMID: 37720749
- PMCID: PMC10500653
- DOI: 10.1021/acsomega.3c03176
Discovery of Novel Naphthoquinone-Chalcone Hybrids as Potent FGFR1 Tyrosine Kinase Inhibitors: Synthesis, Biological Evaluation, and Molecular Modeling
Abstract
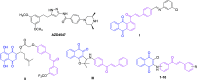
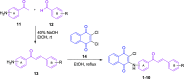
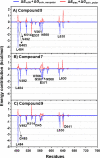
This work presents a flexible synthesis of 10 novel naphthoquinone-chalcone derivatives (1-10) by nucleophilic substitution of readily accessible aminochalcones and 2,3-dichloro-1,4-naphthoquinone. All compounds displayed broad-spectrum cytotoxic activities against all the tested cancer cell lines (i.e., HuCCA-1, HepG2, A549, MOLT-3, T47D, and MDA-MB-231) with IC50 values in the range of 0.81-62.06 μM, especially the four most potent compounds 1, 3, 8, and 9. The in vitro investigation on the fibroblast growth factor receptor 1 (FGFR1) inhibitory effect indicated that eight derivatives (1-2, 4-5, and 7-10) were active FGFR1 inhibitors (IC50 = 0.33-3.13 nM) with more potency than that of the known FGFR1 inhibitor, AZD4547 (IC50 = 12.17 nM). Promisingly, compounds 5 (IC50 = 0.33 ± 0.01 nM), 9 (IC50 = 0.50 ± 0.04 nM), and 7 (IC50 = 0.85 ± 0.08 nM) were the three most potent FGFR1 inhibitors. Molecular docking, molecular dynamics simulations, and MM/GBSA-based free energy calculation revealed that the key amino acid residues involved in the binding of the compounds 5, 7, and 9 and the target FGFR1 protein were similar with those of the AZD4547 (i.e., Val492, Lys514, Ile545, Val561, Ala640, and Asp641). These findings revealed that the newly synthesized naphthoquinone-chalcone scaffold is a promising structural feature for an efficient inhibition of FGFR1.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Discovery of Anilino-1,4-naphthoquinones as Potent EGFR Tyrosine Kinase Inhibitors: Synthesis, Biological Evaluation, and Comprehensive Molecular Modeling.ACS Omega. 2022 May 18;7(21):17881-17893. doi: 10.1021/acsomega.2c01188. eCollection 2022 May 31. ACS Omega. 2022. PMID: 35664590 Free PMC article.
-
Design, synthesis and biological evaluation of novel FGFR inhibitors bearing an indazole scaffold.Org Biomol Chem. 2015 Jul 28;13(28):7643-54. doi: 10.1039/c5ob00778j. Epub 2015 Jun 17. Org Biomol Chem. 2015. PMID: 26080733
-
Design, synthesis and biological evaluation of novel 1H-1,2,4-triazole, benzothiazole and indazole-based derivatives as potent FGFR1 inhibitors viafragment-based virtual screening.J Enzyme Inhib Med Chem. 2020 Dec;35(1):72-84. doi: 10.1080/14756366.2019.1673745. J Enzyme Inhib Med Chem. 2020. PMID: 31682465 Free PMC article.
-
Novel benzothiazole-based dual VEGFR-2/EGFR inhibitors targeting breast and liver cancers: Synthesis, cytotoxic activity, QSAR and molecular docking studies.Bioorg Med Chem Lett. 2022 Feb 15;58:128529. doi: 10.1016/j.bmcl.2022.128529. Epub 2022 Jan 7. Bioorg Med Chem Lett. 2022. PMID: 35007724 Review.
-
Targeting the interplay between MMP-2, CA II and VEGFR-2 via new sulfonamide-tethered isomeric triazole hybrids; Microwave-assisted synthesis, computational studies and evaluation.Bioorg Chem. 2022 Jul;124:105816. doi: 10.1016/j.bioorg.2022.105816. Epub 2022 Apr 16. Bioorg Chem. 2022. PMID: 35489270 Review.
Cited by
-
Synthesis of Benzopyran-Phenylpropanoid Hybrids via Matsuda-Heck-Arylation and Allylic Oxidation.J Org Chem. 2024 Dec 20;89(24):18585-18601. doi: 10.1021/acs.joc.4c02520. Epub 2024 Dec 7. J Org Chem. 2024. PMID: 39644248 Free PMC article.
-
CADD Methods for Developing Novel Compounds Synthesized to Inhibit Tyrosine Kinase Receptors.Curr Top Med Chem. 2025;25(10):1141-1164. doi: 10.2174/0115680266312422240712053821. Curr Top Med Chem. 2025. PMID: 39113295 Review.
References
-
- Carugo A.; Draetta G. F. Academic discovery of anticancer drugs: Historic and future perspectives. Annu. Rev. Cancer Biol. 2018, 3, 385–408. 10.1146/annurev-cancerbio-030518-055645. - DOI
-
- Nass S. J.; Rothenberg M. L.; Pentz R.; Hricak H.; Abernethy A.; Anderson K.; Gee A. W.; Harvey R. D.; Piantadosi S.; Bertagnolli M. M.; Schrag D.; Schilsky R. L. Accelerating anticancer drug development - opportunities and trade-offs. Nat. Rev. Clin. Oncol. 2018, 15, 777–786. 10.1038/s41571-018-0102-3. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

