Cholesterol and Ceramide Facilitate Membrane Fusion Mediated by the Fusion Peptide of the SARS-CoV-2 Spike Protein
- PMID: 37720777
- PMCID: PMC10500581
- DOI: 10.1021/acsomega.3c03610
Cholesterol and Ceramide Facilitate Membrane Fusion Mediated by the Fusion Peptide of the SARS-CoV-2 Spike Protein
Abstract
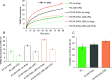
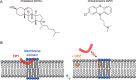
SARS-CoV-2 entry into host cells is mediated by the Spike (S) protein of the viral envelope. The S protein is composed of two subunits: S1 that induces binding to the host cell via its interaction with the ACE2 receptor of the cell surface and S2 that triggers fusion between viral and cellular membranes. Fusion by S2 depends on its heptad repeat domains that bring membranes close together and its fusion peptide (FP) that interacts with and perturbs the membrane structure to trigger fusion. Recent studies have suggested that cholesterol and ceramide lipids from the cell surface may facilitate SARS-CoV-2 entry into host cells, but their exact mode of action remains unknown. We have used a combination of in vitro liposome-liposome and in situ cell-cell fusion assays to study the lipid determinants of S-mediated membrane fusion. Our findings reveal that both cholesterol and ceramide lipids facilitate fusion, suggesting that targeting these lipids could be effective against SARS-CoV-2. As a proof of concept, we examined the effect of chlorpromazine (CPZ), an antipsychotic drug known to perturb membrane structure. Our results show that CPZ effectively inhibits S-mediated membrane fusion, thereby potentially impeding SARS-CoV-2 entry into the host cell.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Kawase M.; Shirato K.; van der Hoek L.; Taguchi F.; Matsuyama S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012, 86, 6537–6545. 10.1128/JVI.00094-12. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

