Finding Lead Compounds for Dengue Antivirals from a Collection of Old Drugs through In Silico Target Prediction and Subsequent In Vitro Validation
- PMID: 37720780
- PMCID: PMC10500654
- DOI: 10.1021/acsomega.3c02607
Finding Lead Compounds for Dengue Antivirals from a Collection of Old Drugs through In Silico Target Prediction and Subsequent In Vitro Validation
Abstract
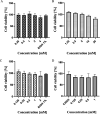
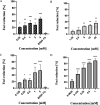
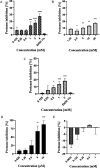
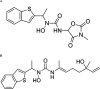
Dengue virus (DENV) infection is one of the most widely spread flavivirus infections. Despite the fatality it could cause, no antiviral treatment is currently available to treat the disease. Hence, this study aimed to repurpose old drugs as novel DENV NS3 inhibitors. Ligand-based (L-B) and proteochemometric (PCM) prediction models were built using 62,354 bioactivity data to screen for potential NS3 inhibitors. Selected drugs were then subjected to the foci forming unit reduction assay (FFURA) and protease inhibition assay. Finally, molecular docking was performed to validate these results. The in silico studies revealed that both models performed well in the internal and external validations. However, the L-B model showed better accuracy in the external validation in terms of its sensitivity (0.671). In the in vitro validation, all drugs (zileuton, trimethadione, and linalool) were able to moderately inhibit the viral activities at the highest concentration tested. Zileuton showed comparable results with linalool when tested at 2 mM against the DENV NS3 protease, with a reduction of protease activity at 17.89 and 18.42%, respectively. Two new compounds were also proposed through the combination of the selected drugs, which are ziltri (zilueton + trimethadione) and zilool (zileuton + linalool). The molecular docking study confirms the in vitro observations where all drugs and proposed compounds were able to achieve binding affinity ≥ -4.1 kcal/mol, with ziltri showing the highest affinity at -7.7 kcal/mol, surpassing the control, panduratin A. The occupation of both S1 and S2 subpockets of NS2B-NS3 may be essential and a reason for the lower binding energy shown by the proposed compounds compared to the screened drugs. Based on the results, this study provided five potential new lead compounds (ziltri, zilool, zileuton, linalool, and trimethadione) for DENV that could be modified further.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
In silico evaluation of inhibitory potential of triterpenoids from Azadirachta indica against therapeutic target of dengue virus, NS2B-NS3 protease.J Vector Borne Dis. 2016 Apr-Jun;53(2):156-61. J Vector Borne Dis. 2016. PMID: 27353586
-
Recent advances in natural products as potential inhibitors of dengue virus with a special emphasis on NS2b/NS3 protease.Phytochemistry. 2022 Oct;202:113362. doi: 10.1016/j.phytochem.2022.113362. Epub 2022 Aug 7. Phytochemistry. 2022. PMID: 35948138 Review.
-
Synthetic peptide optimization improves the inhibition of dengue NS2B-NS3 protease and dengue replication in vitro.Acta Virol. 2019;63(3):278-285. doi: 10.4149/av_2019_305. Acta Virol. 2019. PMID: 31507193
-
Seawater fungi-derived compound screening to identify novel small molecules against dengue virus NS5 methyltransferase and NS2B/NS3 protease.Inform Med Unlocked. 2022;30:100932. doi: 10.1016/j.imu.2022.100932. Epub 2022 Mar 26. Inform Med Unlocked. 2022. PMID: 35372666 Free PMC article.
-
Assessing the potential of NS2B/NS3 protease inhibitors biomarker in curbing dengue virus infections: In silico vs. In vitro approach.Front Cell Infect Microbiol. 2023 Feb 14;13:1061937. doi: 10.3389/fcimb.2023.1061937. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36864886 Free PMC article. Review.
Cited by
-
Machine learning and molecular docking prediction of potential inhibitors against dengue virus.Front Chem. 2024 Dec 24;12:1510029. doi: 10.3389/fchem.2024.1510029. eCollection 2024. Front Chem. 2024. PMID: 39776767 Free PMC article.
-
Novel and repurposed antiviral molecules for arbovirus infections with epidemic Potential: A systematic review.New Microbes New Infect. 2025 Jul 10;66:101614. doi: 10.1016/j.nmni.2025.101614. eCollection 2025 Aug. New Microbes New Infect. 2025. PMID: 40776987 Free PMC article. Review.
-
i-DENV: development of QSAR based regression models for predicting inhibitors targeting non-structural (NS) proteins of dengue virus.Front Pharmacol. 2025 Jun 26;16:1605722. doi: 10.3389/fphar.2025.1605722. eCollection 2025. Front Pharmacol. 2025. PMID: 40642016 Free PMC article.
-
Investigating how dengue virus-induced metabolic changes affect the host immune response and how to develop Immunomodulatory strategies.Virol J. 2025 Apr 25;22(1):117. doi: 10.1186/s12985-025-02745-3. Virol J. 2025. PMID: 40281578 Free PMC article. Review.
References
-
- World Health Organization . Dengue and Severe Dengue, 2018. http://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue. (accessed October 26, 2018).
-
- Dengue Worldwide Overview, 2022. https://www.ecdc.europa.eu/en/dengue-monthly. (accessed July 06, 2022).
-
- First FDA-approved vaccine for the prevention of dengue disease in endemic regions [Press release] U.S. Food and Drug Administration. https://www.fda.gov/news-events/press-announcements/first-fda-approved-v... (accessed July 27, 2023).
LinkOut - more resources
Full Text Sources

