Neural circuit mechanisms for transforming learned olfactory valences into wind-oriented movement
- PMID: 37721371
- PMCID: PMC10588983
- DOI: 10.7554/eLife.85756
Neural circuit mechanisms for transforming learned olfactory valences into wind-oriented movement
Abstract
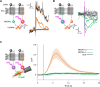
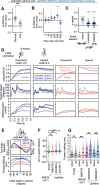
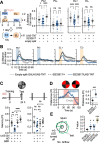
How memories are used by the brain to guide future action is poorly understood. In olfactory associative learning in Drosophila, multiple compartments of the mushroom body act in parallel to assign a valence to a stimulus. Here, we show that appetitive memories stored in different compartments induce different levels of upwind locomotion. Using a photoactivation screen of a new collection of split-GAL4 drivers and EM connectomics, we identified a cluster of neurons postsynaptic to the mushroom body output neurons (MBONs) that can trigger robust upwind steering. These UpWind Neurons (UpWiNs) integrate inhibitory and excitatory synaptic inputs from MBONs of appetitive and aversive memory compartments, respectively. After formation of appetitive memory, UpWiNs acquire enhanced response to reward-predicting odors as the response of the inhibitory presynaptic MBON undergoes depression. Blocking UpWiNs impaired appetitive memory and reduced upwind locomotion during retrieval. Photoactivation of UpWiNs also increased the chance of returning to a location where activation was terminated, suggesting an additional role in olfactory navigation. Thus, our results provide insight into how learned abstract valences are gradually transformed into concrete memory-driven actions through divergent and convergent networks, a neuronal architecture that is commonly found in the vertebrate and invertebrate brains.
Keywords: D. melanogaster; associative learning; dopamine; memory; mushroom body; neural circuit; neuroscience; olfactory navigation.
© 2023, Aso et al.
Conflict of interest statement
YA, DY, DB, KH, MS, HO, YS, TH No competing interests declared
Figures














Update of
References
-
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais PY, Robie AA, Yamagata N, Schnaitmann C, Rowell WJ, Johnston RM, Ngo TTB, Chen N, Korff W, Nitabach MN, Heberlein U, Preat T, Branson KM, Tanimoto H, Rubin GM. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014b;3:e04580. doi: 10.7554/eLife.04580. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

