Lipid-Functionalized Single-Walled Carbon Nanotubes as Probes for Screening Cell Wall Disruptors
- PMID: 37721709
- PMCID: PMC11806933
- DOI: 10.1021/acsami.3c06592
Lipid-Functionalized Single-Walled Carbon Nanotubes as Probes for Screening Cell Wall Disruptors
Abstract
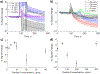
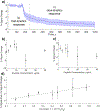
Membrane-active molecules are of great importance to drug delivery and antimicrobials applications. While the ability to prototype new membrane-active molecules has improved greatly with the advent of automated chemistries and rapid biomolecule expression techniques, testing methods are still limited by throughput, cost, and modularity. Existing methods suffer from feasibility constraints of working with pathogenic living cells and by intrinsic limitations of model systems. Herein, we demonstrate an abiotic sensor that uses semiconducting single-walled carbon nanotubes (SWCNTs) as near-infrared fluorescent transducers to report membrane interactions. This sensor is composed of SWCNTs aqueously suspended in lipid, creating a cylindrical, bilayer corona; these SWCNT probes are very sensitive to solvent access (changes in permittivity) and thus report morphological changes to the lipid corona by modulation of fluorescent signals, where binding and disruption are reported as brightening and attenuation, respectively. This mechanism is first demonstrated with chemical and physical membrane-disruptive agents, including ethanol and sodium dodecyl sulfate, and application of electrical pulses. Known cell-penetrating and antimicrobial peptides are then used to demonstrate how the dynamic response of these sensors can be deconvoluted to evaluate different parallel mechanisms of interaction. Last, SWCNTs functionalized in several different bacterial lipopolysaccharides (Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli) are used to evaluate a panel of known membrane-disrupting antimicrobials to demonstrate that drug selectivity can be assessed by suspension of SWCNTs with different membrane materials.
Keywords: cell proxy; disruption; membrane; nanosensor; screening techniques.
Figures
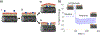
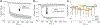

References
-
- Dopp JL; Rothstein SM; Mansell TJ; Reuel NF Rapid Prototyping of Proteins: Mail Order Gene Fragments to Assayable Proteins within 24 Hours. Biotechnol. Bioeng 2019, 116 (3), 667–676. - PubMed
-
- Osterman IA; Komarova ES; Shiryaev DI; Korniltsev IA; Khven IM; Lukyanov DA; Tashlitsky VN; Serebryakova MV; Efremenkova OV; Ivanenkov YA Sorting out Antibiotics’ Mechanisms of Action: A Double Fluorescent Protein Reporter for High-Throughput Screening of Ribosome and DNA Biosynthesis Inhibitors. Antimicrob. Agents Chemother 2016, 60 (12), 7481–7489. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

