GLUT3 promotes macrophage signaling and function via RAS-mediated endocytosis in atopic dermatitis and wound healing
- PMID: 37721853
- PMCID: PMC10617774
- DOI: 10.1172/JCI170706
GLUT3 promotes macrophage signaling and function via RAS-mediated endocytosis in atopic dermatitis and wound healing
Abstract
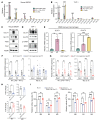
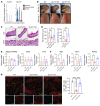
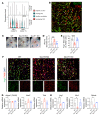
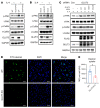
The facilitative GLUT1 and GLUT3 hexose transporters are expressed abundantly in macrophages, but whether they have distinct functions remains unclear. We confirmed that GLUT1 expression increased after M1 polarization stimuli and found that GLUT3 expression increased after M2 stimulation in macrophages. Conditional deletion of Glut3 (LysM-Cre Glut3fl/fl) impaired M2 polarization of bone marrow-derived macrophages. Alternatively activated macrophages from the skin of patients with atopic dermatitis showed increased GLUT3 expression, and a calcipotriol-induced model of atopic dermatitis was rescued in LysM-Cre Glut3fl/fl mice. M2-like macrophages expressed GLUT3 in human wound tissues as assessed by transcriptomics and costaining, and GLUT3 expression was significantly decreased in nonhealing, compared with healing, diabetic foot ulcers. In an excisional wound healing model, LysM-Cre Glut3fl/fl mice showed significantly impaired M2 macrophage polarization and delayed wound healing. GLUT3 promoted IL-4/STAT6 signaling, independently of its glucose transport activity. Unlike plasma membrane-localized GLUT1, GLUT3 was localized primarily to endosomes and was required for the efficient endocytosis of IL-4Rα subunits. GLUT3 interacted directly with GTP-bound RAS in vitro and in vivo through its intracytoplasmic loop domain, and this interaction was required for efficient STAT6 activation and M2 polarization. PAK activation and macropinocytosis were also impaired without GLUT3, suggesting broader roles for GLUT3 in the regulation of endocytosis. Thus, GLUT3 is required for efficient alternative macrophage polarization and function, through a glucose transport-independent, RAS-mediated role in the regulation of endocytosis and IL-4/STAT6 activation.
Keywords: Dermatology; Glucose metabolism; Immunology; Macrophages; Skin.
Figures



Comment in
-
GLUT3 regulates alternative macrophage signaling through a glucose transport-independent role.J Clin Invest. 2023 Nov 1;133(21):e174540. doi: 10.1172/JCI174540. J Clin Invest. 2023. PMID: 37909335 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

