Cholinergic neurons trigger epithelial Ca2+ currents to heal the gut
- PMID: 37722602
- PMCID: PMC10699467
- DOI: 10.1038/s41586-023-06627-y
Cholinergic neurons trigger epithelial Ca2+ currents to heal the gut
Abstract
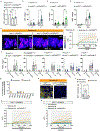
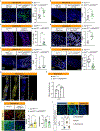
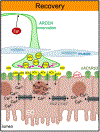
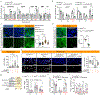
A fundamental and unresolved question in regenerative biology is how tissues return to homeostasis after injury. Answering this question is essential for understanding the aetiology of chronic disorders such as inflammatory bowel diseases and cancer1. We used the Drosophila midgut2 to investigate this and discovered that during regeneration a subpopulation of cholinergic3 neurons triggers Ca2+ currents among intestinal epithelial cells, the enterocytes, to promote return to homeostasis. We found that downregulation of the conserved cholinergic enzyme acetylcholinesterase4 in the gut epithelium enables acetylcholine from specific Egr5 (TNF in mammals)-sensing cholinergic neurons to activate nicotinic receptors in innervated enterocytes. This activation triggers high Ca2+, which spreads in the epithelium through Innexin2-Innexin7 gap junctions6, promoting enterocyte maturation followed by reduction of proliferation and inflammation. Disrupting this process causes chronic injury consisting of ion imbalance, Yki (YAP in humans) activation7, cell death and increase of inflammatory cytokines reminiscent of inflammatory bowel diseases8. Altogether, the conserved cholinergic pathway facilitates epithelial Ca2+ currents that heal the intestinal epithelium. Our findings demonstrate nerve- and bioelectric9-dependent intestinal regeneration and advance our current understanding of how a tissue returns to homeostasis after injury.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures









References
Additional References:
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

