Ecophysiology and interactions of a taurine-respiring bacterium in the mouse gut
- PMID: 37723166
- PMCID: PMC10507020
- DOI: 10.1038/s41467-023-41008-z
Ecophysiology and interactions of a taurine-respiring bacterium in the mouse gut
Abstract
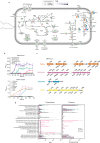
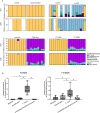
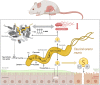
Taurine-respiring gut bacteria produce H2S with ambivalent impact on host health. We report the isolation and ecophysiological characterization of a taurine-respiring mouse gut bacterium. Taurinivorans muris strain LT0009 represents a new widespread species that differs from the human gut sulfidogen Bilophila wadsworthia in its sulfur metabolism pathways and host distribution. T. muris specializes in taurine respiration in vivo, seemingly unaffected by mouse diet and genotype, but is dependent on other bacteria for release of taurine from bile acids. Colonization of T. muris in gnotobiotic mice increased deconjugation of taurine-conjugated bile acids and transcriptional activity of a sulfur metabolism gene-encoding prophage in other commensals, and slightly decreased the abundance of Salmonella enterica, which showed reduced expression of galactonate catabolism genes. Re-analysis of metagenome data from a previous study further suggested that T. muris can contribute to protection against pathogens by the commensal mouse gut microbiota. Together, we show the realized physiological niche of a key murine gut sulfidogen and its interactions with selected gut microbiota members.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

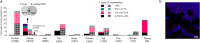

References
-
- Blachier F, et al. Production of hydrogen sulfide by the intestinal microbiota and epithelial cells and consequences for the colonic and rectal mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 2021;320:G125–G135. - PubMed
-
- Blachier F, Beaumont M, Kim E. Cysteine-derived hydrogen sulfide and gut health: a matter of endogenous or bacterial origin. Curr. Opin. Clin. Nutr. Metab. Care. 2019;22:68–75. - PubMed
-
- Attene-Ramos MS, Wagner ED, Rex Gaskins H, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 2007;5:455–459. - PubMed
-
- Babidge W, Millard S, Roediger W. Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: implications for ulcerative colitis. Mol. Cell. Biochem. 1998;181:117–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

