Statistically unbiased prediction enables accurate denoising of voltage imaging data
- PMID: 37723246
- PMCID: PMC10555843
- DOI: 10.1038/s41592-023-02005-8
Statistically unbiased prediction enables accurate denoising of voltage imaging data
Abstract
Here we report SUPPORT (statistically unbiased prediction utilizing spatiotemporal information in imaging data), a self-supervised learning method for removing Poisson-Gaussian noise in voltage imaging data. SUPPORT is based on the insight that a pixel value in voltage imaging data is highly dependent on its spatiotemporal neighboring pixels, even when its temporally adjacent frames alone do not provide useful information for statistical prediction. Such dependency is captured and used by a convolutional neural network with a spatiotemporal blind spot to accurately denoise voltage imaging data in which the existence of the action potential in a time frame cannot be inferred by the information in other frames. Through simulations and experiments, we show that SUPPORT enables precise denoising of voltage imaging data and other types of microscopy image while preserving the underlying dynamics within the scene.
© 2023. The Author(s).
Conflict of interest statement
Y.-G.Y., M.E. and S.H. declare the following competing interests. Y.-G.Y., M.E. and S.H. are co-inventors on patent applications owned by KAIST covering SUPPORT (KR10-2023-0091724).
Figures

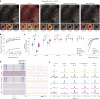
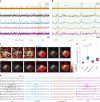


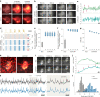


References
-
- Yoon YG, et al. Sparse decomposition light-field microscopy for high speed imaging of neuronal activity. Optica. 2020;7:1457–1468. doi: 10.1364/OPTICA.392805. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

