Longitudinal genomic surveillance of carriage and transmission of Clostridioides difficile in an intensive care unit
- PMID: 37723252
- PMCID: PMC10579090
- DOI: 10.1038/s41591-023-02549-4
Longitudinal genomic surveillance of carriage and transmission of Clostridioides difficile in an intensive care unit
Abstract
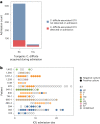
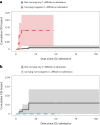
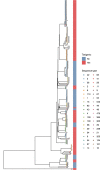
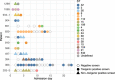
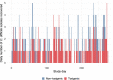
Despite enhanced infection prevention efforts, Clostridioides difficile remains the leading cause of healthcare-associated infections in the United States. Current prevention strategies are limited by their failure to account for patients who carry C. difficile asymptomatically, who may act as hidden reservoirs transmitting infections to other patients. To improve the understanding of asymptomatic carriers' contribution to C. difficile spread, we conducted admission and daily longitudinal culture-based screening for C. difficile in a US-based intensive care unit over nine months and performed whole-genome sequencing on all recovered isolates. Despite a high burden of carriage, with 9.3% of admissions having toxigenic C. difficile detected in at least one sample, only 1% of patients culturing negative on admission to the unit acquired C. difficile via cross-transmission. While patients who carried toxigenic C. difficile on admission posed minimal risk to others, they themselves had a 24-times greater risk for developing a healthcare-onset C. difficile infection than noncarriers. Together, these findings suggest that current infection prevention practices can be effective in preventing nosocomial cross-transmission of C. difficile, and that decreasing C. difficile infections in hospitals further will require interventions targeting the transition from asymptomatic carriage to infection.
© 2023. The Author(s).
Conflict of interest statement
V.B.Y. has consulted for Vedanta Biosciences, and has received support via a Debiopharm grant. He is a mSphere senior editor. He sits on the Advisory Council of the University of Oklahoma Health Sciences Center COBRE. The other authors declare no competing interests.
Figures



Comment in
-
Genomic epidemiology of Clostridioides difficile in individuals admitted to an intensive care unit.Nat Med. 2023 Oct;29(10):2418-2419. doi: 10.1038/s41591-023-02550-x. Nat Med. 2023. PMID: 37783971 No abstract available.
References
-
- Antibiotic Resistance Threats in the United States (Centers for Disease Control and Prevention, 2019); 10.15620/cdc:82532
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

