Frequency of change determines effectiveness of microbial response strategies
- PMID: 37723339
- PMCID: PMC10579261
- DOI: 10.1038/s41396-023-01515-9
Frequency of change determines effectiveness of microbial response strategies
Abstract
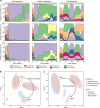
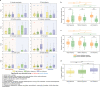
Nature challenges microbes with change at different frequencies and demands an effective response for survival. Here, we used controlled laboratory experiments to investigate the effectiveness of different response strategies, such as post-translational modification, transcriptional regulation, and specialized versus adaptable metabolisms. For this, we inoculated replicated chemostats with an enrichment culture obtained from sulfidic stream microbiomes 16 weeks prior. The chemostats were submitted to alternatingly oxic and anoxic conditions at three frequencies, with periods of 1, 4 and 16 days. The microbial response was recorded with 16S rRNA gene amplicon sequencing, shotgun metagenomics, transcriptomics and proteomics. Metagenomics resolved provisional genomes of all abundant bacterial populations, mainly affiliated with Proteobacteria and Bacteroidetes. Almost all these populations maintained a steady growth rate under both redox conditions at all three frequencies of change. Our results supported three conclusions: (1) Oscillating oxic/anoxic conditions selected for generalistic species, rather than species specializing in only a single condition. (2) A high frequency of change selected for strong codon usage bias. (3) Alignment of transcriptomes and proteomes required multiple generations and was dependent on a low frequency of change.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–29. - PubMed
-
- Mitsui A, Kumazawa S, Takahashi A, Ikemoto H, Cao S, Arai T. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–2.
-
- Golden SS, Canales SR. Cyanobacterial circadian clocks—timing is everything. Nat Rev Microbiol. 2003;1:191–9. - PubMed

