Characterization of 29 newly isolated bacteriophages as a potential therapeutic agent against IMP-6-producing Klebsiella pneumoniae from clinical specimens
- PMID: 37724861
- PMCID: PMC10581060
- DOI: 10.1128/spectrum.04761-22
Characterization of 29 newly isolated bacteriophages as a potential therapeutic agent against IMP-6-producing Klebsiella pneumoniae from clinical specimens
Abstract
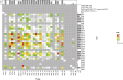
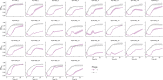
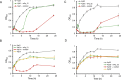
Carbapenemase-producing Enterobacteriaceae (CPE) are one of the most detrimental species of antibiotic-resistant bacteria globally. Phage therapy has emerged as an effective strategy for the treatment of CPE infections. In western Japan, the rise of Klebsiella pneumoniae strains harboring the pKPI-6 plasmid encoding bla IMP-6 is of increasing concern. To address this challenge, we isolated 29 phages from Japanese sewage, specifically targeting 31 K. pneumoniae strains and one Escherichia coli strain harboring the pKPI-6 plasmid. Electron microscopy analysis revealed that among the 29 isolated phages, 21 (72.4%), 5 (17.2%), and 3 (10.3%) phages belonged to myovirus, siphovirus, and podovirus morphotypes, respectively. Host range analysis showed that 18 Slopekvirus strains within the isolated phages infected 25-26 K. pneumoniae strains, indicating that most of the isolated phages have a broad host range. Notably, K. pneumoniae strain Kp21 was exclusively susceptible to phage øKp_21, whereas Kp22 exhibited susceptibility to over 20 phages. Upon administering a phage cocktail composed of 10 phages, we observed delayed emergence of phage-resistant bacteria in Kp21 but not in Kp22. Intriguingly, phage-resistant Kp21 exhibited heightened sensitivity to other bacteriophages, indicating a "trade-off" for resistance to phage øKp_21. Our proposed phage set has an adequate number of phages to combat the K. pneumoniae strain prevalent in Japan, underscoring the potential of a well-designed phage cocktail in mitigating the occurrence of phage-resistant bacteria. IMPORTANCE The emergence of Klebsiella pneumoniae harboring the bla IMP-6 plasmid poses an escalating threat in Japan. In this study, we found 29 newly isolated bacteriophages that infect K. pneumoniae strains carrying the pKPI-6 plasmid from clinical settings in western Japan. Our phages exhibited a broad host range. We applied a phage cocktail treatment composed of 10 phages against two host strains, Kp21 and Kp22, which displayed varying phage susceptibility patterns. Although the phage cocktail delayed the emergence of phage-resistant Kp21, it was unable to hinder the emergence of phage-resistant Kp22. Moreover, the phage-resistant Kp21 became sensitive to other phages that were originally non-infective to the wild-type Kp21 strains. Our study highlights the potential of a well-tailored phage cocktail in reducing the occurrence of phage-resistant bacteria.
Keywords: Klebsiella pneumoniae; bacteriophages; bla IMP-6; phage library; phage-resistant bacteria; trade-off.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Yamagishi T, Matsui M, Sekizuka T, Ito H, Fukusumi M, Uehira T, Tsubokura M, Ogawa Y, Miyamoto A, Nakamori S, Tawa A, Yoshimura T, Yoshida H, Hirokawa H, Suzuki S, Matsui T, Shibayama K, Kuroda M, Oishi K. 2020. A prolonged multispecies outbreak of IMP-6 carbapenemase-producing enterobacterales due to horizontal transmission of the IncN plasmid. Sci Rep 10:4139. doi: 10.1038/s41598-020-60659-2 - DOI - PMC - PubMed
-
- Abe R, Akeda Y, Sugawara Y, Takeuchi D, Matsumoto Y, Motooka D, Yamamoto N, Kawahara R, Tomono K, Fujino Y, Hamada S, Langelier C. 2020. Characterization of the plasmidome encoding carbapenemase and mechanisms for dissemination of carbapenem-resistant Enterobacteriaceae. mSystems 5:e00759-20. doi: 10.1128/mSystems.00759-20 - DOI - PMC - PubMed
-
- Kayama S, Shigemoto N, Kuwahara R, Oshima K, Hirakawa H, Hisatsune J, Jové T, Nishio H, Yamasaki K, Wada Y, Ueshimo T, Miura T, Sueda T, Onodera M, Yokozaki M, Hattori M, Ohge H, Sugai M. 2015. Complete nucleotide sequence of the IncN plasmid encoding Imp-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother 59:1356–1359. doi: 10.1128/AAC.04759-14 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

