Evolution of the T4 phage virion is driven by selection pressure from non-bacterial factors
- PMID: 37724862
- PMCID: PMC10580926
- DOI: 10.1128/spectrum.00115-23
Evolution of the T4 phage virion is driven by selection pressure from non-bacterial factors
Abstract
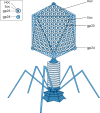
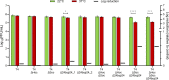
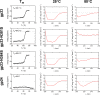
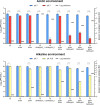
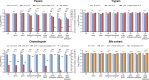
Bacteriophages colonize animal and human bodies, propagating on sensitive bacteria that are symbionts, commensals, or pathogens of animals and humans. T4-like phages are dependent on abundant symbionts such as Escherichia coli, commonly present in animal and human gastrointestinal (GI) tracts. Bacteriophage T4 is one of the most complex viruses, and its intricate structure, particularly the capsid head protecting the phage genome, likely contributes substantially to the overall phage fitness in diverse environments. We investigated how individual head proteins-gp24, Hoc, and Soc-affect T4 phage survival under pressure from non-bacterial factors. We constructed a panel of T4 phage variants defective in these structural proteins: T4∆Soc, T4∆24byp24, T4∆Hoc∆Soc, T4∆Hoc∆24byp24, T4∆Soc∆24byp24, and T4∆Hoc∆Soc∆24byp24 (byp = bypass). These variants were investigated for their sensitivity to selected environmental conditions relevant to the microenvironment of the GI tract, including pH, temperature, and digestive enzymes. The simple and "primitive" structure of the phage capsid (∆24byp24) was significantly less stable at low pH and more sensitive to inactivation by digestive enzymes, and the simultaneous lack of gp24 and Soc resulted in a notable decrease in phage activity at 37°C. Gp24 was also found to be highly resistant to thermal and chemical denaturation. Thus, gp24, which was acquired relatively late in evolution, seems to play a key role in T4 withstanding environmental conditions, including those related to the animal/human GI tract, and Soc is a molecular glue that enhances this protective effect. IMPORTANCE Bacteriophages are important components of animal and human microbiota, particularly in the gastrointestinal tract, where they dominate the viral community and contribute to shaping microbial balance. However, interactions with bacterial hosts are not the only element of the equation in phage survival-phages inhabiting the GI tract are constantly exposed to increased temperature, pH fluctuations, or digestive enzymes, which raises the question of whether and how the complex structure of phage capsids contributes to their persistence in the specific microenvironment of human/animal bodies. Here we address this phage-centric perspective, identifying the role of individual head proteins in T4 phage survival in GI tract conditions. The selection pressure driving the evolution of T4-like phages could have come from the external environment that affects phage virions with increased temperature and variable pH; it is possible that in the local microenvironment along the GI tract, the phage benefits from stability-protecting proteins.
Keywords: T4 phage; bacteriophage evolution; gastrointestinal tract; gp24; head proteins; head vertex protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Modulation of bacteriophage T4 capsid size.Virology. 1996 Jul 1;221(1):67-77. doi: 10.1006/viro.1996.0353. Virology. 1996. PMID: 8661415
-
Immunogenicity studies of proteins forming the T4 phage head surface.J Virol. 2014 Nov;88(21):12551-7. doi: 10.1128/JVI.02043-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142581 Free PMC article.
-
Structure and assembly of bacteriophage T4 head.Virol J. 2010 Dec 3;7:356. doi: 10.1186/1743-422X-7-356. Virol J. 2010. PMID: 21129201 Free PMC article. Review.
-
Recombinant expression and purification of T4 phage Hoc, Soc, gp23, gp24 proteins in native conformations with stability studies.PLoS One. 2012;7(7):e38902. doi: 10.1371/journal.pone.0038902. Epub 2012 Jul 13. PLoS One. 2012. PMID: 22808021 Free PMC article.
-
Genomes of the T4-related bacteriophages as windows on microbial genome evolution.Virol J. 2010 Oct 28;7:292. doi: 10.1186/1743-422X-7-292. Virol J. 2010. PMID: 21029436 Free PMC article. Review.
Cited by
-
Super-Resolution Goes Viral: T4 Virus Particles as Versatile 3D-Bio-NanoRulers.Adv Mater. 2025 Mar;37(12):e2403365. doi: 10.1002/adma.202403365. Epub 2025 Jan 16. Adv Mater. 2025. PMID: 39821930 Free PMC article.
-
Advances and optimization strategies in bacteriophage therapy for treating inflammatory bowel disease.Front Immunol. 2024 May 8;15:1398652. doi: 10.3389/fimmu.2024.1398652. eCollection 2024. Front Immunol. 2024. PMID: 38779682 Free PMC article. Review.
-
Episymbiotic Saccharibacteria TM7x modulates the susceptibility of its host bacteria to phage infection and promotes their coexistence.Proc Natl Acad Sci U S A. 2024 Apr 16;121(16):e2319790121. doi: 10.1073/pnas.2319790121. Epub 2024 Apr 9. Proc Natl Acad Sci U S A. 2024. PMID: 38593079 Free PMC article.
References
LinkOut - more resources
Full Text Sources

