Severe pneumonia and pathogenic damage in human airway epithelium caused by Coxsackievirus B4
- PMID: 37725516
- PMCID: PMC10538465
- DOI: 10.1080/22221751.2023.2261560
Severe pneumonia and pathogenic damage in human airway epithelium caused by Coxsackievirus B4
Abstract
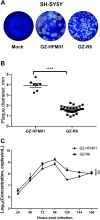
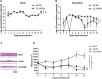
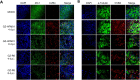
Coxsackievirus B4 (CVB4) has one of the highest proportions of fatal outcomes of other enterovirus serotypes. However, the pathogenesis of severe respiratory disease caused by CVB4 infection remains unclear. In this study, 3 of 42 (7.2%, GZ-R6, GZ-R7 and GZ-R8) patients with severe pneumonia tested positive for CVB4 infection in southern China. Three full-length genomes of pneumonia-derived CVB4 were sequenced and annotated for the first time, showing their high nucleotide similarity and clustering within genotype V. To analyze the pathogenic damage caused by CVB4 in the lungs, a well-differentiated human airway epithelium (HAE) was established and infected with the pneumonia-derived CVB4 isolate GZ-R6. The outcome was compared with that of a severe hand-foot-mouth disease (HFMD)-derived CVB4 strain GZ-HFM01. Compared with HFMD-derived CVB4, pneumonia-derived CVB4 caused more intense and rapid disruption of HAE polarity, leading to tight-junction barrier disruption, loss of cilia, and airway epithelial cell hypertrophy. More pneumonia-derived CVB4 were released from the basolateral side of the HAE than HFMD-derived CVB4. Of the 18 cytokines tested, only IL-6 and IL-1b secretion significantly increased on bilateral sides of HAE during the early stage of pneumonia-derived CVB4 infection, while multiple cytokine secretions significantly increased in HFMD-derived CVB4-infected HAE. HFMD-derived CVB4 exhibited stronger neurovirulence in the human neuroblastoma cells SH-SY5Y than pneumonia-derived CVB4, which is consistent with the clinical manifestations of patients infected with these two viruses. This study has increased the depth of our knowledge of severe pneumonia infection caused by CVB4 and will benefit its prevention and treatment.
Keywords: Coxsackievirus B4; full-length genome; hand-foot-mouth disease (HFMD); human airway epithelium; neurovirulence; pathogenic damage; severe pneumonia.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
New coxsackievirus B4 genotype circulating in Inner Mongolia Autonomous Region, China.PLoS One. 2014 Mar 3;9(3):e90379. doi: 10.1371/journal.pone.0090379. eCollection 2014. PLoS One. 2014. PMID: 24595311 Free PMC article.
-
Genomic Epidemiology and Transmission Dynamics of Global Coxsackievirus B4.Viruses. 2023 Feb 19;15(2):569. doi: 10.3390/v15020569. Viruses. 2023. PMID: 36851788 Free PMC article.
-
Complete genome sequence of a recombinant coxsackievirus B4 from a patient with a fatal case of hand, foot, and mouth disease in Guangxi, China.J Virol. 2012 Oct;86(19):10901-2. doi: 10.1128/JVI.01808-12. J Virol. 2012. PMID: 22966193 Free PMC article.
-
Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide.Expert Rev Anti Infect Ther. 2015;13(9):1061-71. doi: 10.1586/14787210.2015.1058156. Epub 2015 Jun 25. Expert Rev Anti Infect Ther. 2015. PMID: 26112307 Review.
-
Atypical hand-foot-mouth disease in Belém, Amazon region, northern Brazil, with detection of coxsackievirus A6.J Clin Virol. 2020 May;126:104307. doi: 10.1016/j.jcv.2020.104307. Epub 2020 Mar 6. J Clin Virol. 2020. PMID: 32302950 Review.
Cited by
-
Epidemiological and genetic characterizations of hand, foot, and mouth disease and acute respiratory infections due to CV-A6 infection in Henan Province, China between 2021 and 2022.BMC Pediatr. 2025 Apr 2;25(1):264. doi: 10.1186/s12887-025-05527-6. BMC Pediatr. 2025. PMID: 40170027 Free PMC article.
-
The multiple roles of viral 3Dpol protein in picornavirus infections.Virulence. 2024 Dec;15(1):2333562. doi: 10.1080/21505594.2024.2333562. Epub 2024 Apr 15. Virulence. 2024. PMID: 38622757 Free PMC article. Review.
-
Pseudorabies virus infection increases the permeability of the mammalian respiratory barrier to facilitate Pasteurella multocida infection.mSphere. 2024 Aug 28;9(8):e0029724. doi: 10.1128/msphere.00297-24. Epub 2024 Jul 23. mSphere. 2024. PMID: 39041808 Free PMC article.
-
Immunogenicity and protective efficacy of inactivated coxsackievirus B4 viral particles.Emerg Microbes Infect. 2024 Dec;13(1):2337665. doi: 10.1080/22221751.2024.2337665. Epub 2024 Apr 5. Emerg Microbes Infect. 2024. PMID: 38551145 Free PMC article.
References
-
- Crowell RL, Landau BJ.. A short history and introductory background on the coxsackieviruses of group B. Curr Top Microbiol Immunol. 1997;223:1–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
