Structure of pre-miR-31 reveals an active role in Dicer-TRBP complex processing
- PMID: 37725636
- PMCID: PMC10523476
- DOI: 10.1073/pnas.2300527120
Structure of pre-miR-31 reveals an active role in Dicer-TRBP complex processing
Abstract
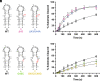
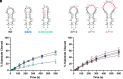
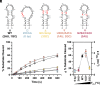
As an essential posttranscriptional regulator of gene expression, microRNA (miRNA) levels must be strictly maintained. The biogenesis of many miRNAs is mediated by trans-acting protein partners through a variety of mechanisms, including remodeling of the RNA structure. miR-31 functions as an oncogene in numerous cancers, and interestingly, its biogenesis is not known to be regulated by protein-binding partners. Therefore, the intrinsic structural properties of the precursor element of miR-31 (pre-miR-31) can provide a mechanism by which its biogenesis is regulated. We determined the solution structure of pre-miR-31 to investigate the role of distinct structural elements in regulating processing by the Dicer-TRBP complex. We found that the presence or absence of mismatches within the helical stem does not strongly influence Dicer-TRBP processing of the pre-miRNAs. However, both the apical loop size and structure at the Dicing site are key elements for discrimination by the Dicer-TRBP complex. Interestingly, our NMR-derived structure reveals the presence of a triplet of base pairs that link the Dicer cleavage site and the apical loop. Mutational analysis in this region suggests that the stability of the junction region strongly influences processing by the Dicer-TRBP complex. Our results enrich our understanding of the active role that RNA structure plays in regulating miRNA biogenesis, which has direct implications for the control of gene expression.
Keywords: NMR spectroscopy; RNA; microRNA; structural biology.
Conflict of interest statement
The authors declare no competing interest.
Figures



Update of
-
Structure of pre-miR-31 reveals an active role in Dicer processing.bioRxiv [Preprint]. 2023 Jan 3:2023.01.03.519659. doi: 10.1101/2023.01.03.519659. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2300527120. doi: 10.1073/pnas.2300527120. PMID: 36711709 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

