Disassembly of bundled F-actin and cellular remodeling via an interplay of Mical, cofilin, and F-actin crosslinkers
- PMID: 37725655
- PMCID: PMC10523612
- DOI: 10.1073/pnas.2309955120
Disassembly of bundled F-actin and cellular remodeling via an interplay of Mical, cofilin, and F-actin crosslinkers
Abstract
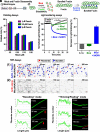
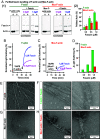
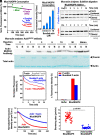
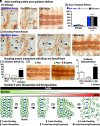
Cellular form and function are controlled by the assembly and stability of actin cytoskeletal structures-but disassembling/pruning these structures is equally essential for the plasticity and remodeling that underlie behavioral adaptations. Importantly, the mechanisms of actin assembly have been well-defined-including that it is driven by actin's polymerization into filaments (F-actin) and then often bundling by crosslinking proteins into stable higher-order structures. In contrast, it remains less clear how these stable bundled F-actin structures are rapidly disassembled. We now uncover mechanisms that rapidly and extensively disassemble bundled F-actin. Using biochemical, structural, and imaging assays with purified proteins, we show that F-actin bundled with one of the most prominent crosslinkers, fascin, is extensively disassembled by Mical, the F-actin disassembly enzyme. Furthermore, the product of this Mical effect, Mical-oxidized actin, is poorly bundled by fascin, thereby further amplifying Mical's disassembly effects on bundled F-actin. Moreover, another critical F-actin regulator, cofilin, also affects fascin-bundled filaments, but we find herein that it synergizes with Mical to dramatically amplify its disassembly of bundled F-actin compared to the sum of their individual effects. Genetic and high-resolution cellular assays reveal that Mical also counteracts crosslinking proteins/bundled F-actin in vivo to control cellular extension, axon guidance, and Semaphorin/Plexin cell-cell repulsion. Yet, our results also support the idea that fascin-bundling serves to dampen Mical's F-actin disassembly in vitro and in vivo-and that physiologically relevant cellular remodeling requires a fine-tuned interplay between the factors that build bundled F-actin networks and those that disassemble them.
Keywords: MICAL1; MICAL2; MICAL3; bristle; nervous system.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
F-actin dismantling through a redox-driven synergy between Mical and cofilin.Nat Cell Biol. 2016 Aug;18(8):876-85. doi: 10.1038/ncb3390. Epub 2016 Jul 25. Nat Cell Biol. 2016. PMID: 27454820 Free PMC article.
-
Profilin and Mical combine to impair F-actin assembly and promote disassembly and remodeling.Nat Commun. 2021 Sep 20;12(1):5542. doi: 10.1038/s41467-021-25781-3. Nat Commun. 2021. PMID: 34545088 Free PMC article.
-
The MICALs are a Family of F-actin Dismantling Oxidoreductases Conserved from Drosophila to Humans.Sci Rep. 2018 Jan 17;8(1):937. doi: 10.1038/s41598-017-17943-5. Sci Rep. 2018. PMID: 29343822 Free PMC article.
-
MICAL-mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics.Front Cell Dev Biol. 2023 Feb 17;11:1124202. doi: 10.3389/fcell.2023.1124202. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36875759 Free PMC article. Review.
-
Extracellular inhibitors, repellents, and semaphorin/plexin/MICAL-mediated actin filament disassembly.Cytoskeleton (Hoboken). 2011 Aug;68(8):415-33. doi: 10.1002/cm.20527. Epub 2011 Aug 25. Cytoskeleton (Hoboken). 2011. PMID: 21800438 Free PMC article. Review.
Cited by
-
Autoinhibition and relief mechanisms for MICAL monooxygenases in F-actin disassembly.Nat Commun. 2024 Aug 9;15(1):6824. doi: 10.1038/s41467-024-50940-7. Nat Commun. 2024. PMID: 39122694 Free PMC article.
-
Actin network evolution as a key driver of eukaryotic diversification.J Cell Sci. 2024 Aug 1;137(15):jcs261660. doi: 10.1242/jcs.261660. Epub 2024 Aug 9. J Cell Sci. 2024. PMID: 39120594 Free PMC article. Review.
-
The Cdc42/Rac1 pathway: a molecular mechanism behind iron-deficiency-driven aortic medial degeneration.Am J Transl Res. 2024 Aug 15;16(8):3922-3937. doi: 10.62347/YISX1726. eCollection 2024. Am J Transl Res. 2024. PMID: 39262709 Free PMC article.
-
Chemokine Receptor Antagonists Prevent and Reverse Cofilin-Actin Rod Pathology and Protect Synapses in Cultured Rodent and Human iPSC-Derived Neurons.Biomedicines. 2024 Jan 1;12(1):93. doi: 10.3390/biomedicines12010093. Biomedicines. 2024. PMID: 38255199 Free PMC article.
-
HIV-1 budding requires cortical actin disassembly by the oxidoreductase MICAL1.Proc Natl Acad Sci U S A. 2024 Nov 26;121(48):e2407835121. doi: 10.1073/pnas.2407835121. Epub 2024 Nov 18. Proc Natl Acad Sci U S A. 2024. PMID: 39556735 Free PMC article.
References
-
- Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014). - PubMed
-
- Miyoshi T., Watanabe N., Can filament treadmilling alone account for the F-actin turnover in lamellipodia? Cytoskeleton (Hoboken) 70, 179–190 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

