Correlative CD4 and CD8 T-cell immunodominance in humans and mice: Implications for preclinical testing
- PMID: 37726420
- PMCID: PMC10616275
- DOI: 10.1038/s41423-023-01083-0
Correlative CD4 and CD8 T-cell immunodominance in humans and mice: Implications for preclinical testing
Erratum in
-
Correction: Correlative CD4 and CD8 T-cell immunodominance in humans and mice: implications for preclinical testing.Cell Mol Immunol. 2023 Dec;20(12):1534. doi: 10.1038/s41423-023-01099-6. Cell Mol Immunol. 2023. PMID: 37964123 Free PMC article. No abstract available.
Abstract
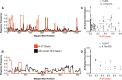
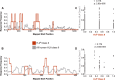
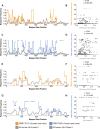
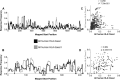
Antigen-specific T-cell recognition is restricted by Major Histocompatibility Complex (MHC) molecules, and differences between CD4 and CD8 immunogenicity in humans and animal species used in preclinical vaccine testing are yet to be fully understood. In this study, we addressed this matter by analyzing experimentally identified epitopes based on published data curated in the Immune Epitopes DataBase (IEDB) database. We first analyzed SARS-CoV-2 spike (S) and nucleoprotein (N), which are two common targets of the immune response and well studied in both human and mouse systems. We observed a weak but statistically significant correlation between human and H-2b mouse T-cell responses (CD8 S specific (r = 0.206, p = 1.37 × 10-13); CD4 S specific (r = 0.118, p = 2.63 × 10-5) and N specific (r = 0.179, p = 2.55 × 10-4)). Due to intrinsic differences in MHC molecules across species, we also investigated the association between the immunodominance of common Human Leukocyte Antigen (HLA) alleles for which HLA transgenic mice are available, namely, A*02:01, B*07:02, DRB1*01:01, and DRB1*04:01, and found higher significant correlations for both CD8 and CD4 (maximum r = 0.702, p = 1.36 × 10-31 and r = 0.594, p = 3.04-122, respectively). Our results further indicated that some regions are commonly immunogenic between humans and mice (either H-2b or HLA transgenic) but that others are human specific. Finally, we noted a significant correlation between CD8 and CD4 S- (r = 0.258, p = 7.33 × 1021) and N-specific (r = 0.369, p = 2.43 × 1014) responses, suggesting that discrete protein subregions can be simultaneously recognized by T cells. These findings were confirmed in other viral systems, providing general guidance for the use of murine models to test T-cell immunogenicity of viral antigens destined for human use.
Keywords: Animal testing; HLA; SARS-CoV-2; T cells; Vaccine.
© 2023. The Author(s), under exclusive licence to CSI and USTC.
Conflict of interest statement
AS is a consultant for AstraZeneca Pharmaceuticals, Calyptus Pharmaceuticals, Inc, Darwin Health, EmerVax, EUROIMMUN, F. Hoffman-La Roche Ltd, Fortress Biotech, Gilead Sciences, Granite bio., Gritstone Oncology, Guggenheim Securities, Moderna, Pfizer, RiverVest Venture Partners, and Turnstone Biologics. AG is a consultant for Pfizer. LJI has filed for patent protection for various aspects of T-cell epitope and vaccine design work. JS and TAPN declare no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

