Development of an automated, high-throughput SARS-CoV-2 neutralization assay based on a pseudotyped virus using a vesicular stomatitis virus (VSV) vector
- PMID: 37727107
- PMCID: PMC10540657
- DOI: 10.1080/22221751.2023.2261566
Development of an automated, high-throughput SARS-CoV-2 neutralization assay based on a pseudotyped virus using a vesicular stomatitis virus (VSV) vector
Abstract
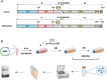
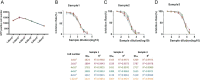
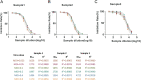
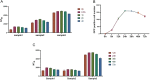
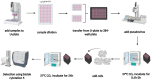
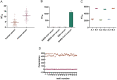
ABSTRACTThe global outbreak of COVID-19 has caused a severe threat to human health; therefore, simple, high-throughput neutralization assays are desirable for developing vaccines and drugs against COVID-19. In this study, a high-titre SARS-CoV-2 pseudovirus was successfully packaged by truncating the C-terminus of the SARS-CoV-2 spike protein by 21 amino acids and infecting 293 T cells that had been stably transfected with the angiotensin-converting enzyme 2 (ACE2) receptor and furin (named AF cells), to establish a simple, high-throughput, and automated 384-well plate neutralization assay. The method was optimized for cell amount, virus inoculation, incubation time, and detection time. The automated assay showed good sensitivity, accuracy, reproducibility, Z' factor, and a good correlation with the live virus neutralization assay. The high-throughput approach would make it available for the SARS-CoV-2 neutralization test in large-scale clinical trials and seroepidemiological surveys which would aid the accelerated vaccine development and evaluation.
Keywords: SARS-CoV-2; VSV; high-throughput; neutralization assay; pseudovirus.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
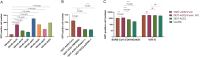

Similar articles
-
Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells.Emerg Microbes Infect. 2020 Dec;9(1):2105-2113. doi: 10.1080/22221751.2020.1815589. Emerg Microbes Infect. 2020. PMID: 32893735 Free PMC article.
-
Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein.J Virol. 2020 Oct 14;94(21):e01062-20. doi: 10.1128/JVI.01062-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32788194 Free PMC article.
-
Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein.Virol J. 2021 Jan 12;18(1):16. doi: 10.1186/s12985-021-01490-7. Virol J. 2021. PMID: 33435994 Free PMC article.
-
Pseudotyped Vesicular Stomatitis Virus-Severe Acute Respiratory Syndrome-Coronavirus-2 Spike for the Study of Variants, Vaccines, and Therapeutics Against Coronavirus Disease 2019.Front Microbiol. 2022 Jan 14;12:817200. doi: 10.3389/fmicb.2021.817200. eCollection 2021. Front Microbiol. 2022. PMID: 35095820 Free PMC article. Review.
-
SARS-CoV-2 Neutralization Assays Used in Clinical Trials: A Narrative Review.Vaccines (Basel). 2024 May 18;12(5):554. doi: 10.3390/vaccines12050554. Vaccines (Basel). 2024. PMID: 38793805 Free PMC article. Review.
Cited by
-
Construction of pseudotyped human coronaviruses and detection of pre-existing antibodies in the human population.Biosaf Health. 2024 Sep 3;6(5):279-285. doi: 10.1016/j.bsheal.2024.09.002. eCollection 2024 Oct. Biosaf Health. 2024. PMID: 40078732 Free PMC article.
-
Pseudotyped Viruses: A Useful Platform for Pre-Clinical Studies Conducted in a BSL-2 Laboratory Setting.Biomolecules. 2025 Jan 15;15(1):135. doi: 10.3390/biom15010135. Biomolecules. 2025. PMID: 39858529 Free PMC article. Review.
References
-
- Aleem A, Akbar Samad AB, Vaqar S. Emerging variants of SARS-CoV-2 and novel therapeutics against Coronavirus (COVID-19). StatPearls. Treasure Island (FL), 2023. - PubMed
-
- Chan JF, Kok KH, Zhu Z, et al. . Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020 Dec;9(1):221–236. doi:10.1080/22221751.2020.1719902. PubMed PMID: 31987001; PubMed Central PMCID: PMCPMC7067204. - DOI - PMC - PubMed
-
- Scholer L, Le-Trilling VTK, Eilbrecht M, et al. . A novel in-cell ELISA assay allows rapid and automated quantification of SARS-CoV-2 to analyze neutralizing antibodies and antiviral compounds. Front Immunol. 2020;11:573526, doi:10.3389/fimmu.2020.573526. PubMed PMID: 33162987; PubMed Central PMCID: PMCPMC7581787. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
