Transcriptome profiling of intact bowel wall reveals that PDE1A and SEMA3D are possible markers with roles in enteric smooth muscle apoptosis, proliferative disorders, and dysautonomia in Crohn's disease
- PMID: 37727374
- PMCID: PMC10505932
- DOI: 10.3389/fgene.2023.1194882
Transcriptome profiling of intact bowel wall reveals that PDE1A and SEMA3D are possible markers with roles in enteric smooth muscle apoptosis, proliferative disorders, and dysautonomia in Crohn's disease
Abstract
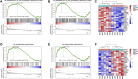
Background: Inflammatory bowel disease (IBD) is a complex and multifactorial inflammatory condition, comprising Crohn's disease (CD) and ulcerative colitis (UC). While numerous studies have explored the immune response in IBD through transcriptional profiling of the enteric mucosa, the subtle distinctions in the pathogenesis of Crohn's disease and ulcerative colitis remain insufficiently understood. Methods: The intact bowel wall specimens from IBD surgical patients were divided based on their inflammatory status into inflamed Crohn's disease (iCD), inflamed ulcerative colitis (iUC) and non-inflamed (niBD) groups for RNA sequencing. Differential mRNA GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes), and GSEA (Gene Set Enrichment Analysis) bioinformatic analyses were performed with a focus on the enteric autonomic nervous system (ANS) and smooth muscle cell (SMC). The transcriptome results were validated by quantitative polymerase chain reaction (qPCR) and immunohistochemistry (IHC). Results: A total of 2099 differentially expressed genes were identified from the comparison between iCD and iUC. Regulation of SMC apoptosis and proliferation were significantly enriched in iCD, but not in iUC. The involved gene PDE1A in iCD was 4-fold and 1.5-fold upregulated at qPCR and IHC compared to that in iUC. Moreover, only iCD was significantly associated with the gene sets of ANS abnormality. The involved gene SEMA3D in iCD was upregulated 8- and 5-fold at qPCR and IHC levels compared to iUC. Conclusion: These findings suggest that PDE1A and SEMA3D may serve as potential markers implicated in enteric smooth muscle apoptosis, proliferative disorders, and dysautonomia specifically in Crohn's disease.
Keywords: Crohn’s disease; PDE1A; RNA seq; Sema3D; apoptosis; autonomic nervous system; proliferation; smooth muscle cell.
Copyright © 2023 Yang, Xia, Yang, Wang, Meng, Zhang, Ma, Gou, Wang, Shu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ahrens R., Waddell A., Seidu L., Blanchard C., Carey R., Forbes E., et al. (2008). Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J. Immunol. 181 (10), 7390–7399. 10.4049/jimmunol.181.10.7390 - DOI - PMC - PubMed
-
- Anders S., Huber W. (2012). Differential expression of RNA-Seq data at the gene level – the DESeq package.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

