AP003352.1/miR-141-3p axis enhances the proliferation of osteosarcoma by LPAR3
- PMID: 37727685
- PMCID: PMC10506581
- DOI: 10.7717/peerj.15937
AP003352.1/miR-141-3p axis enhances the proliferation of osteosarcoma by LPAR3
Abstract
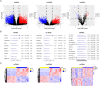
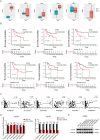
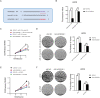
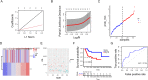
Osteosarcoma (OS) is a highly malignant tumor with a poor prognosis and a growing incidence. LncRNAs and microRNAs control the occurrence and development process of osteosarcoma through ceRNA patterns. The LPAR3 gene is important in cancer cell proliferation, apoptosis and disease development. However, the regulatory mechanism of the ceRNA network through which LPAR3 participates in osteosarcoma has not been clarified. Herein, our study demonstrated that the AP003352.1/miR-141-3p axis drives LPAR3 expression to induce the malignant progression of osteosarcoma. First, the expression of LPAR3 is regulated by the changes in AP003352.1 and miR-141-3p. Similar to the ceRNA of miR-141-3p, AP003352.1 regulates the expression of LPAR3 through this mechanism. In addition, the regulation of AP003352.1 in malignant osteosarcoma progression depends to a certain degree on miR-141-3p. Importantly, the AP003352.1/miR-141-3p/LPAR3 axis can better serve as a multi-gene diagnostic marker for osteosarcoma. In conclusion, our research reveals a new ceRNA regulatory network, which provides a novel potential target for the diagnosis and treatment of osteosarcoma.
Keywords: CeRNA; Cell proliferation; LPAR3; Osteosarcoma.
© 2023 Yu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Long Noncoding RNA HCG9 Promotes Osteosarcoma Progression through RAD51 by Acting as a ceRNA of miR-34b-3p.Mediators Inflamm. 2021 Aug 18;2021:9978882. doi: 10.1155/2021/9978882. eCollection 2021. Mediators Inflamm. 2021. PMID: 34456631 Free PMC article.
-
LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis.Biomed Pharmacother. 2018 Jan;97:1645-1653. doi: 10.1016/j.biopha.2017.12.004. Epub 2017 Dec 8. Biomed Pharmacother. 2018. PMID: 29793327
-
EBF1-mediated up-regulation of lncRNA FGD5-AS1 facilitates osteosarcoma progression by regulating miR-124-3p/G3BP2 axis as a ceRNA.J Orthop Surg Res. 2022 Jun 27;17(1):332. doi: 10.1186/s13018-022-03181-7. J Orthop Surg Res. 2022. PMID: 35761386 Free PMC article.
-
Long noncoding RNA SNHG14 promotes osteosarcoma progression via miR-433-3p/FBXO22 axis.Biochem Biophys Res Commun. 2020 Mar 12;523(3):766-772. doi: 10.1016/j.bbrc.2020.01.016. Epub 2020 Jan 14. Biochem Biophys Res Commun. 2020. PMID: 31948764
-
MiR-455-3p downregulation facilitates cell proliferation and invasion and predicts poor prognosis of osteosarcoma.J Orthop Surg Res. 2020 Oct 2;15(1):454. doi: 10.1186/s13018-020-01967-1. J Orthop Surg Res. 2020. PMID: 33008443 Free PMC article.
Cited by
-
Osteosarcoma in a ceRNET perspective.J Biomed Sci. 2024 Jun 5;31(1):59. doi: 10.1186/s12929-024-01049-y. J Biomed Sci. 2024. PMID: 38835012 Free PMC article. Review.
-
Single-cell RNA and bulk sequencing analysis reveals that formononetin inhibits GTSF1 to exert anti-osteosarcoma effects.APL Bioeng. 2025 Aug 8;9(3):036110. doi: 10.1063/5.0266780. eCollection 2025 Sep. APL Bioeng. 2025. PMID: 40786706 Free PMC article.
References
-
- Aure MR, Fleischer T, Bjorklund S, Ankill J, Castro-Mondragon JA, Osbreac Borresen-Dale AL, Tost J, Sahlberg KK, Mathelier A, Tekpli X, Kristensen VN. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Medicine. 2021;13(1):72. doi: 10.1186/s13073-021-00880-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

