DEVOUR: Deleterious Variants on Uncovered Regions in Whole-Exome Sequencing
- PMID: 37727687
- PMCID: PMC10506587
- DOI: 10.7717/peerj.16026
DEVOUR: Deleterious Variants on Uncovered Regions in Whole-Exome Sequencing
Abstract
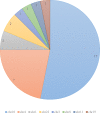
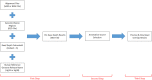
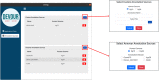
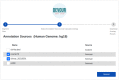
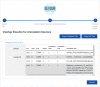
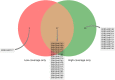
The discovery of low-coverage (i.e. uncovered) regions containing clinically significant variants, especially when they are related to the patient's clinical phenotype, is critical for whole-exome sequencing (WES) based clinical diagnosis. Therefore, it is essential to develop tools to identify the existence of clinically important variants in low-coverage regions. Here, we introduce a desktop application, namely DEVOUR (DEleterious Variants On Uncovered Regions), that analyzes read alignments for WES experiments, identifies genomic regions with no or low-coverage (read depth < 5) and then annotates known variants in the low-coverage regions using clinical variant annotation databases. As a proof of concept, DEVOUR was used to analyze a total of 28 samples from a publicly available Hirschsprung disease-related WES project (NCBI Bioproject: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB19327), revealing the potential existence of 98 disease-associated variants in low-coverage regions. DEVOUR is available from https://github.com/projectDevour/DEVOUR under the MIT license.
Keywords: Clinical NGS informatics; Genetic diseases; Genetic disposition to disease; Genetic variants; Medical genetics; Next-generation sequence (NGS) analysis; Whole-exome sequencing (WES) analysis.
©2023 Türk et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Bergant G, Maver A, Lovrecic L, Cuturilo G, Hodzic A, Peterlin B. Comprehensive use of extended exome analysis improves diagnostic yield in rare disease: a retrospective survey in 1,059 cases. Genetics in Medicine. 2018;20:303–312. - PubMed
-
- Bick AG, Flannick J, Ito K, Cheng S, Vasan RS, Parfenov MG, Herman DS, De Palma SR, Gupta N, Gabriel SB, Funke BH, Rehm HL, Benjamin EJ, Aragam J, Taylor J, Herman A. Fox ER, Newton-Cheh C, Kathiresan S, O’Donnell CJ, Wilson JG, Altshuler DM, Hirschhorn JN, Seidman JG, Seidman C. Burden of rare sarcomere gene variants in the Framingham and Jackson heart study Cohorts. American Journal of Human Genetics. 2012;91:513. doi: 10.1016/j.ajhg.2012.07.017. - DOI - PMC - PubMed
-
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Özen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19096. doi: 10.1073/pnas.0910672106. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

